Modelling
Contents |
Mathematical modelling: introduction
Mathematical modelling plays nowadays a central role in Synthetic Biology, due to its ability to serve as a crucial link between the concept and realization of a biological circuit: what we propose in this page is a modelling approach to our project, which has proven extremely useful and very helpful before and after the "wet lab".
Thus, immediately at the beginning, when there was little knowledge, a mathematical model based on a system of differential equations was derived and implemented using a set of parameters, to validate the feasibility of the project. Once this became clear, starting from the characterization of the single subparts created in the wet lab, some of the parameters of the mathematical model were estimated (the others are known from literature) and they have been fixed to simulate the same model, in order to predict the final behaviour of the whole engineered closed-loop circuit. This approach is consistent with the typical one adopted for the analysis and synthesis of a biological circuit, as exemplified by Pasotti et al 2011.
Therefore here, after a brief overview about the advantages that modelling engineered circuits can bring, we deeply analyze the system of equation formulas, underlining the role and function of the parameters involved.
Experimental procedures for parameter estimation are discussed and, finally, a different type of circuit is presented. Simulations were performed, using ODEs with MATLAB and used to explain the difference between a closed-loop control system model and an open one.
The importance of mathematical modelling
Mathematical modeling reveals fundamental in the challenge for understanding and engineering complex biological systems. Indeed, these are characterized by a high degree of interconnection among the single constituent parts, requiring a comprehensive analysis of their behavior through mathematical formalisms and computational tools
- Simulation: mathematical models allow to analyse complex system dynamics and to reveal the relationships between the involved variables, starting from the knowledge of the behavior of single subparts and from simple hypotesis of their interconnection. (Endler et al, 2009)
- Knowledge elicitation: mathematical models summarize into a small set of parameters the results of several experiments (parameter identification), allowing a robust comparison among different experimental conditions and providing an efficient way to synthesize knowledge about biological processes. Then, through the simulation process, they make possible the re-usability of the knowledge coming from different experiments, engineering complex systems from the composition of its constituent subparts under appropriate experimental/environmental conditions (Braun et al, 2005; Canton et al, 2008)..
Equations for gene networks
 |


Hypothesis of the model
|
HP1: In order to better investigate the range of dynamics of each subparts, every promoter has been
studied with 4 different RBSs, so as to develop more knowledge about the state variables in several configurations
of RBS' efficiency (Salis et al, 2009). Hereafter, referring to the notation "RBSx" we mean, respectively,
RBS30,
RBS31,
RBS32,
RBS34.
HP2: In equation (2) only HSL is considered as inducer, instead of the complex LuxR-HSL. This is motivated by the fact that our final device offers a constitutive LuxR production due to the upstream constitutive promoter P&lambda. Assuming LuxR is abundant and always saturated in the cytoplasm, we can justify the simplification of attributing pLux promoter i nduction only by HSL. In conclusion LuxR, LuxI and AiiA were not included in the equation system. HP3: in system equation, LuxI and AiiA amounts are expressed per cell. For this reason, the whole equation (3), except for the term of intrinsic degradation of HSL, is multiplied by the number of cells N, due to the property of the lactone to diffuse freely inside/outside bacteria. HP4: as regards promoters pTet and pLux, we assume their strengths (measured in PoPs), due to a given concentration of inducer (aTc, HSL for Ptet and Plux respectively), to be independent from the gene encoding. In other words, in our hypothesis, if the mRFP coding region is substituted with a region coding for another gene (in our case, AiiA or LuxI), we would obtain the same synthesis rate: this is the reason why the strength of the complex promoter-RBSx is expressed in Arbitrary Units [AUr]. HP5: considering the exponential growth, the enzymes AiiA and LuxI concentration is supposed to be constant, because their production is equally compensated by dilution. |
Equations (1) and (2)
Equations (1) and (2) have identical structure, differing only in the parameters involved. They represent the synthesis degradation and diluition of both the enzymes in the circuit, LuxI and AiiA, respectively in the first and second equation: in each of them both transcription and translation processes have been condensed. The corresponding mathematical formalism is analogous to the one used by Pasotti et al 2011, Suppl. Inf., even if we do not take LuxR-HSL complex formation into account, as explained below.
These equations are composed of 2 parts:
The first term describes, through Hill's equation formalism, the synthesis rate of the protein of interest (either LuxI or AiiA) depending on the concentration of the inducer (anhydrotetracicline -aTc- or HSL respectively), responsible for the activation of the regulatory element composed of promoter and RBSx. In the parameter table (see below), α refers to the maximum activation of the promoter, while δ stands for its leakage activity (this means that the promoter is slightly active even if there is no induction). In particular, in equation (1), the almost entire inhibition of pTet promoter is given by the constitutive production of TetR by our MGZ1 strain. In equation (2), pLux is almost inactive in the absence of the complex LuxR-HSL.
Furthermore, in both equations k stands for the dissociation constant of the promoter from the inducer (respectively aTc and HSL in eq. 1 and 2), while η is the cooperativity constant.
The second term in equations (1) and (2) is in turn composed of 2 parts. The former one (γ*LuxI or γ*AiiA respectively) describes, with an exponential decay, the degradation rate per cell of the protein. The latter (μ*(Nmax-N)/Nmax)*LuxI or μ*(Nmax-N)/Nmax)*AiiA, respectively) takes into account the dilution factor against cell growth which is related to the cell replication process.
Equation (3)
Here the kinetics of HSL is modeled, through enzymatic reactions either related to the production or the degradation of HSL: based on the experiments performed, we derived appropriate expressions for HSL synthesis and degradation. This equation is composed of 3 parts:
- The first term represents the production of HSL due to LuxI expression. We modeled this process with a saturation curve in which Vmax is the HSL maximum transcription rate, while kM,LuxI is the dissociation constant of LuxI from the substrate HSL.
- The second term represents the degradation of HSL due to the AiiA expression. Similarly to LuxI, kcat represents the maximum degradation per unit of HSL concentration, while kM,AiiA is the concentration at which AiiA dependent HSL concentration rate is (kcat*HSL)/2. The formalism is similar to that found in the Supplementary Information of Danino et al, 2010.
- The third term (γHSL*HSL) is similar to the corresponding ones present in the first two equations and describes the intrinsic protein degradation.
Equation (4)
Table of parameters and species
| Parameter & Species | Description | Unit of Measurement | Value |
| αpTet | maximum transcription rate of pTet (related to RBSx efficiency) | [(AUr/min)/cell] | - |
| δpTet | leakage factor of promoter pTet basic activity | [-] | - |
| ηpTet | Hill coefficient of pTet | [-] | - |
| kpTet | dissociation constant of aTc from pTet | [nM] | - |
| αpLux | maximum transcription rate of pLux (related to RBSx efficiency) | [(AUr/min)/cell] | - |
| δpLux | leakage factor of promoter pLux basic activity | [-] | - |
| ηpLux | Hill coefficient of pLux | [-] | - |
| kpLux | dissociation constant of HSL from pLux | [nM] | - |
| γpLux | LuxI constant degradation | [1/min] | - |
| γAiiA | AiiA constant degradation | [1/min] | - |
| γHSL | HSL constant degradation | [1/min] | - |
| Vmax | maximum transcription rate of LuxI | [nM/(min*cell)] | - |
| kM,LuxI | dissociation constant of LuxI from HSL | [AUr/cell] | - |
| kcat | maximum number of enzymatic reactions catalyzed per minute | [1/(min*cell)] | - |
| kM,AiiA | dissociation constant of AiiA from HSL | [AUr/cell] | - |
| Nmax | maximum number of bacteria per well | [cell] | - |
| μ | rate of bacteria growth | [1/min] | - |
| LuxI | kinetics of enzyme LuxI | [AUr⁄cell] | - |
| AiiA | kinetics of enzyme AiiA | [AUr⁄cell] | - |
| HSL | kinetics of HSL | [nM⁄(min)] | - |
| N | number of cells | cell | - |
Parameter CV
| Parameter & Species | BBa_B0030 | BBa_B0031 | BBa_B0032 | BBa_B0034 |
| αpTet | 2.13 | imm | 10.9 | 12.2 |
| δpTet | 888.5 | 1.47*10^11 | 411.3 | 61.74 |
| ηpTet | 12 | imm | 76671474 | 2705172 |
| kpTet | 2.54 | imm | 393180 | 55889 |
| αpLux | 10.14 | 7.13 | 2.78 | 5.8 |
| δpLux | 179.7 | 57.04 | 1317.7 | 187.2 |
| ηpLux | 47.73 | 29.13 | 9.75 | 19.3 |
| kpLux | 27.5 | 25.81 | 8.46 | 17.86 |
Parameter estimation
Promoter (PTet & pLux)
 |
 |
As shown in the figure below, we considered a range of inductions and we monitored, in time, absorbance (O.D. stands for "optical density") and fluorescence; the two vertical segments for each graph highlight the exponential phase of bacterial growth. Scell (namely, synthesis rate per cell) can be derived as a function of inducer concentration, thereby providing the desired input-output relation (inducer concentration versus promoter+RBS activity), which was modelled as a Hill curve:
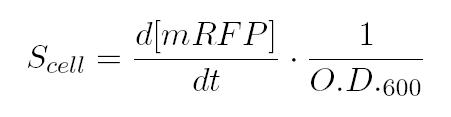

T9002 introduction
|
LuxI and AiiA tests have been always performed exploiting the well-characterized BioBrick BBa_T9002, by which it's possible to quantify exactly the concentration of HSL.
 |
AiiA & LuxI
 |
 |


N
Degradation rates
Simulations
In order to see that, we implemented and simulated in Matlab an open loop circuit, similar to CTRL+E, except for the constitutive production of AiiA.
 |
 |
Sensitivity Analysis of the steady state of enzyme expression in exponential phase
In this paragraph we want to theoretically investigate our circuit behaviour in cell's culture exponential growth phase. According to this, we first derive, under feasible hypoteses, the steady state condition for our enzymes concentration. Then we perform a sensitivity analysis relating the output of our system (HSL) to input (atc) and system parameters.
Steady state of enzyme expression
References
-
Braun D, Basu S, Weiss R (2005) Parameter estimation for two synthetic gene networks: a case study
ICASSP '05 5:v/769-v/772.
Canton B, Labno A, Endy D. (2008) Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 26(7):787-93.
Danino T, Mondragón-Palomino O, Tsimring L et al. (2010) A synchronized quorum of genetic clocks. Nature. 463(7279):326-30.
Endler L, Rodriguez N, Juty N et al. (2009) Designing and encoding models for synthetic biology. J. R. Soc. Interface 6:S405-S417.
Kelly JR, Rubin AJ, Davis JH et al. (2009) Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol. Eng. 3:4.
Pasotti L, Quattrocelli M, Galli D et al. (2011) Multiplexing and demultiplexing logic functions for computing signal processing tasks in synthetic biology. Biotechnol. J. 6(7):784-95.
Salis H M, Mirsky E A, Voight C A (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol.27:946-950.
 "
"