Team:Harvard/Project/Zinc Finger Background
From 2011.igem.org
Overview | Design | Synthesize | Test | Zinc Finger Background | Protocols
What are Zinc Finger Proteins (ZFPs)?
An interview with Dr. George Church and Dr. Keith Joung, discussing zinc fingers and their own research:
Function
ZFPs are found commonly in nature as a class of special transcription factors that bind to DNA, thus regulating gene expression. Zinc finger function was first studied using zinc finger protein Zif268. They became of interest to researchers because of their ability to bind to specific DNA sequences. Since proteins, such as nucleases, can be attatched to a zinc finger protein, scientists could use zinc fingers to interact with, and even modify, the genome at very specific locations.
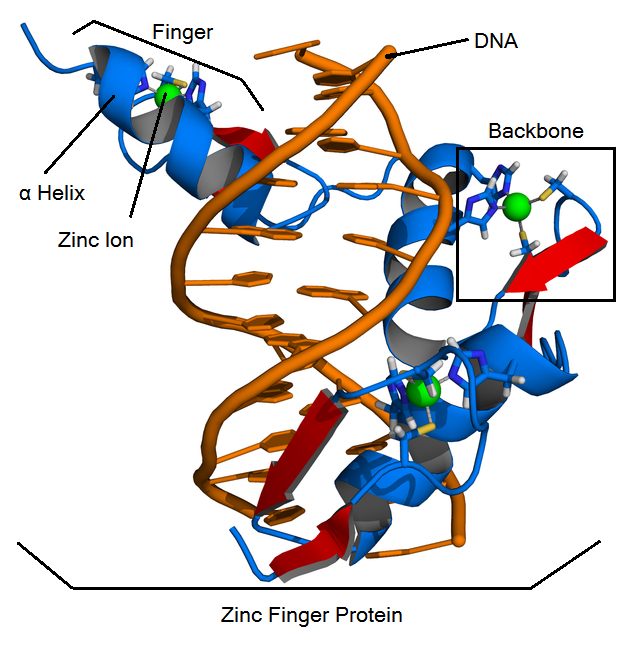
Structure
ZFPs consist of smaller subunits called "fingers" which each contain a zinc finger binding helix that binds to unique DNA sequences. These fingers are linear and linked together by the "zinc finger backbone", a series of approximately 21 amino acids.
Cis2His2 ZFPs have three main structural components:
- Zinc finger binding helix
- Linker region
- Zinc ion that is coordinated by two cysteine residues and two histidine residues.
Terminology
- Backbone: contains most of the amino acids of a zinc finger protein: zif268 is the most famous backbone.
- Fingers: contain a backbone and a helix, bind to a 3-base DNA triplet
- Helix: the alpha helix in a finger. It is responsible for binding to a DNA triplet. Helices are made up of 7 amino acids, and fit into a specified position in a backbone. Amino acid positions are specified by -1,1,2,3,4,5, and 6.
- Zinc finger proteins (ZFPs): arrays of three fingers that bind to 9 bases (3 triplets) of DNA.
Zinc Finger Binding
Current research suggests that the helix region of a zinc finger plays the primary role in determining which DNA triplet it will bind to. Scientists have designed a binding model (pictured to the right), that predicts which amino acid in the helix interacts with which DNA base.
Multiple zinc fingers can be connected with linkers to bind to longer sequences of DNA. However, zinc finger-DNA binding is not completely modular. If zinc finger 1 binds to GGC and zinc finger 2 binds to TTC, one cannot assume that an array composed of these two proteins will bind to GGC-TTC. This is due to an additional interaction between the helix and the nucleotide 1 base upstream from the triplet.
Helpful Zinc Finger Links
[http://en.wikipedia.org/wiki/Zinc_finger Zinc Fingers on Wikipedia]
- A more detailed introduction to zinc fingers.
[http://compbio.cs.princeton.edu/zf/ Predicting DNA Recognition by C2H2 Zinc Finger Proteins]
- A program useful for predicting how well a given amino acid sequence will bind to a given DNA sequence
[http://www.zincfingers.org/default2.htm The Zinc Finger Consortium]
- Information & helpful resources for zinc fingers
[http://www.jounglab.org/ Joung Lab]
- Information about Dr. Joung's extensive work with zinc fingers
 "
"