July, 4th
Digestions of previously purified plasmids were performed for ligations:
| Plasmid |
Kind |
DNA (μl) |
H2O (μl) |
Enzyme 1 (μl) |
Enzyme 2 (μl) |
Buffer H (μl) |
Final Volume (μl) |
| <partinfo>BBa_C0060</partinfo> |
Insert |
16.5 |
4 |
1 EcoRI |
1 SpeI |
2.5 |
25 |
| <partinfo>BBa_C0061</partinfo> |
Insert |
13.2 |
7.3 |
1 XbaI |
1 PstI |
2.5 |
25 |
| <partinfo>BBa_K081022</partinfo> |
Insert |
15.7 |
4.8 |
1 EcoRI |
1 PstI |
2.5 |
25 |
| <partinfo>BBa_B0030</partinfo> |
Vector |
13.2 |
7.3 |
1 SpeI |
1 PstI |
2.5 |
25 |
| <partinfo>BBa_B0031</partinfo> |
Vector |
12.4 |
8.1 |
1 SpeI |
1 PstI |
2.5 |
25 |
| <partinfo>BBa_B0032</partinfo> |
Vector |
9.5 |
11 |
1 SpeI |
1 PstI |
2.5 |
25 |
| <partinfo>BBa_B0015</partinfo> |
Vector |
7.9 |
12.6 |
1 EcoRI |
1 XbaI |
2.5 |
25 |
| <partinfo>pSB4C5</partinfo> |
Vector |
3 |
17.5 |
1 EcoRI |
1 PstI |
2.5 |
25 |
| <partinfo>BBa_I13501</partinfo> |
Insert |
7.2 |
13.3 |
1 XbaI |
1 PstI |
2.5 |
25 |
Reactions were incubated at 37°C for three hours while a small-size and a medium-size agarose gel were prepared according to protocols.
In the afternoon gel electrophoresis was performed.
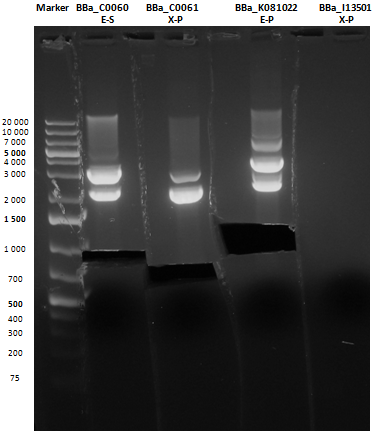
Small size gel
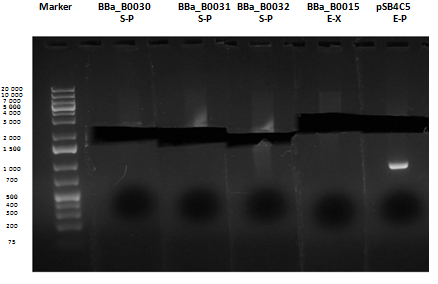
Medium size gel
As shown, in figure all clones were positive (apart from <partinfo>BBa_I13501</partinfo> run where we inexplicably didn't see any band), so we cut and purified the bands of interest. Cells harbouring <partinfo>BBa_I13501</partinfo> were again inoculated in 5 ml LB + Amp.
After gel extraction, cut DNA was quantified:
| Plasmid |
DNA (ng/μl) |
| <partinfo>BBa_C0060</partinfo> (E-S) |
3.9 |
| <partinfo>BBa_C0061</partinfo> (X-P) |
4.1 |
| <partinfo>BBa_K081022</partinfo> (E-P) |
4.4 |
| <partinfo>BBa_B0030</partinfo> (S-P) |
10.3 |
| <partinfo>BBa_B0031</partinfo> (S-P) |
11.8 |
| <partinfo>BBa_B0032</partinfo> (S-P) |
10.1 |
| <partinfo>BBa_B0015</partinfo> (E-X) |
12 |
| <partinfo>pSB4C5</partinfo> (E-P) |
10.7 |
Then ligations were performed in a final volume of 10 μl:
| Ligation Name |
Vector |
Vector volume (μl) |
Insert |
Insert volume (μl) |
Buffer (μl) |
T4 Ligase (μl) |
| E1 |
<partinfo>BBa_B0015</partinfo> (E-X) |
1.5 |
<partinfo>BBa_C0060</partinfo> (E-S) |
6.5 |
1 |
1 |
| E2 |
<partinfo>BBa_B0030</partinfo> (S-P) |
1.5 |
<partinfo>BBa_C0061</partinfo> (X-P) |
6.5 |
1 |
1 |
| E3 |
<partinfo>BBa_B0031</partinfo> (S-P) |
1.5 |
<partinfo>BBa_C0061</partinfo> (X-P) |
6.5 |
1 |
1 |
| E4 |
<partinfo>BBa_B0032</partinfo> (S-P) |
1.5 |
<partinfo>BBa_C0061</partinfo> (X-P) |
6.5 |
1 |
1 |
| E8 |
<partinfo>pSB4C5</partinfo> (E-P) |
1.5 |
<partinfo>BBa_K081022</partinfo> (E-P) |
6.5 |
1 |
1 |
Ligations were incubated ON at 16°C.
July, 5th
T4 Ligase was inactivated, heating ligations at 65°C for 10 minutes; ligations were stored at -20°C. Plasmid purification was done again for <partinfo>BBa_I13501</partinfo> and purified DNA was quantified:
| Plasmid |
DNA (ng/μl) |
| <partinfo>BBa_I13501</partinfo> |
97.2 |
<partinfo>BBa_I13501</partinfo> was then digested with restriction endonucleases:
| Plasmid |
Kind |
DNA (μl) |
H2O (μl) |
Enzyme 1 (μl) |
Enzyme 2 (μl) |
Buffer H (μl) |
Final Volume (μl) |
| <partinfo>BBa_I13501</partinfo> |
Insert |
10.5 |
10 |
1 XbaI |
1 PstI |
2.5 |
25 |
According to protocols the reaction was incubated at 37°C for 3 hours.
60 ml of 80% glycerol were prepared mixing 48 ml of 100% glycerol with 12 ml of ddH2O.
Gel electrophoresis was done for <partinfo>BBa_I13501</partinfo> digestion:

Small size gel
Cut DNA was gel-extracted:
| Plasmid |
DNA (ng/μl) |
| <partinfo>BBa_I13501</partinfo> (X-P) |
2.6 |
Further ligations were prepared and incubated ON at 16°C:
| Ligation Name |
Vector |
Vector volume (μl) |
Insert |
Insert volume (μl) |
Buffer (μl) |
T4 Ligase (μl) |
| E5 |
<partinfo>BBa_B0030</partinfo> (S-P) |
1 |
<partinfo>BBa_I13501</partinfo> (X-P) |
7 |
1 |
1 |
| E6 |
<partinfo>BBa_B0031</partinfo> (S-P) |
1 |
<partinfo>BBa_I13501</partinfo> (X-P) |
7 |
1 |
1 |
| E7 |
<partinfo>BBa_B0032</partinfo> (S-P) |
1 |
<partinfo>BBa_I13501</partinfo> (X-P) |
7 |
1 |
1 |
July, 6th
<partinfo>BBa_C0261</partinfo> was resuspended from iGEM 2011 kit distribution, Plate 1, well 14C in 15 μl of ddH2O; <partinfo>BBa_C0261</partinfo>, E1, E2, E3, E4, E5, E6, E7, and E8 were transformed in 100 μl of TOP10 competent cells according to protocols. Plates were incubated ON at 37°C.
500 ml of LB without antibiotic were prepared.
July, 7th
All plates showed a lot of colonies, except for E-8 (5 colonies); two colonies for E1, E2, E3, E4, E5, E6, E7 plates and three colonies for E8 plate were picked and inoculated in 10 μl LB for inoculum and screening PCR.
A 20x mix was prepared for PCR reaction:
| H2O (μl) |
Buffer 10x (μl) |
MgCl2 (μl) |
VF2 (<partinfo>BBa_G00100</partinfo>) μl |
VR (<partinfo>BBa_G00101</partinfo>) μl |
dNTPs (μl) |
Taq polymerase (μl) |
| 360 |
50 |
20 |
10 |
10 |
10 |
20 |
24 μl were aliquoted in each tube, together with 1 μl of each liquid culture. PCR cycles parameters were set as follows:
- 94°C 30 seconds (denaturing)
- 60°C 1 minute (annealing)
- 72°C 1 minute and 20 seconds (elongation)
35 cycles were performed.
Liquid cultures were then inoculated in 800 μl LB with the proper antibiotic.
A small size and a medium size agarose gel were prepared.
After the PCR, gel run was carried out on the samples:
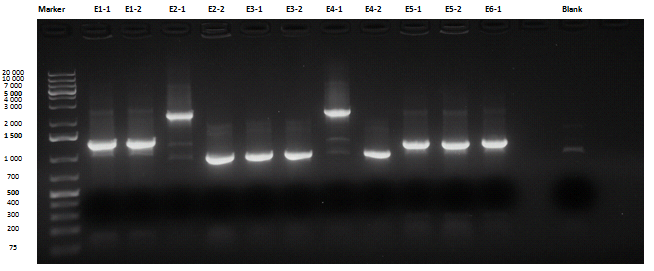
Medium size gel
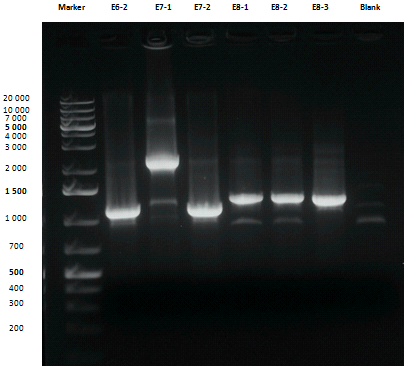
Small size gel
Except for E2-1, E4-1 and E7-1, other band lengths were correct; 750 μl of E1-2, E2-2, E3-1, E4-2, E5-2, E6-1, E7-2 and E8-3 liquid cultures were used to prepare glycerol stocks while the remaining 50 μl were refilled to 5 ml for plasmid purification and incubated at 37°C 220 rpm. <partinfo>BBa_R0040</partinfo>, <partinfo>BBa_C0261</partinfo> and <partinfo>BBa_I13521</partinfo> were also inoculated.
In the late afternoon the team met to talk about the wet-lab activity, the abstract and the bio-safety section.
July, 8th
Cultures were saturated; plasmid purification was carried out:
| Plasmid |
DNA (ng/μl) |
| E1-2 |
64.1 |
| E2-2 |
56.2 |
| E3-1 |
53.5 |
| E4-2 |
67.2 |
| E5-2 |
73.5 |
| E6-1 |
92.2 |
| E7-2 |
67.6 |
| E8-3 |
25.3 |
| <partinfo>BBa_R0040</partinfo> |
48.1 |
| <partinfo>BBa_C0261</partinfo> |
61.2 |
| <partinfo>BBa_I13521</partinfo> |
79.3 |
 "
"




