Team:DTU-Denmark/Project
From 2011.igem.org
(→Abstract) |
(→Abstract) |
||
| Line 3: | Line 3: | ||
== Abstract == | == Abstract == | ||
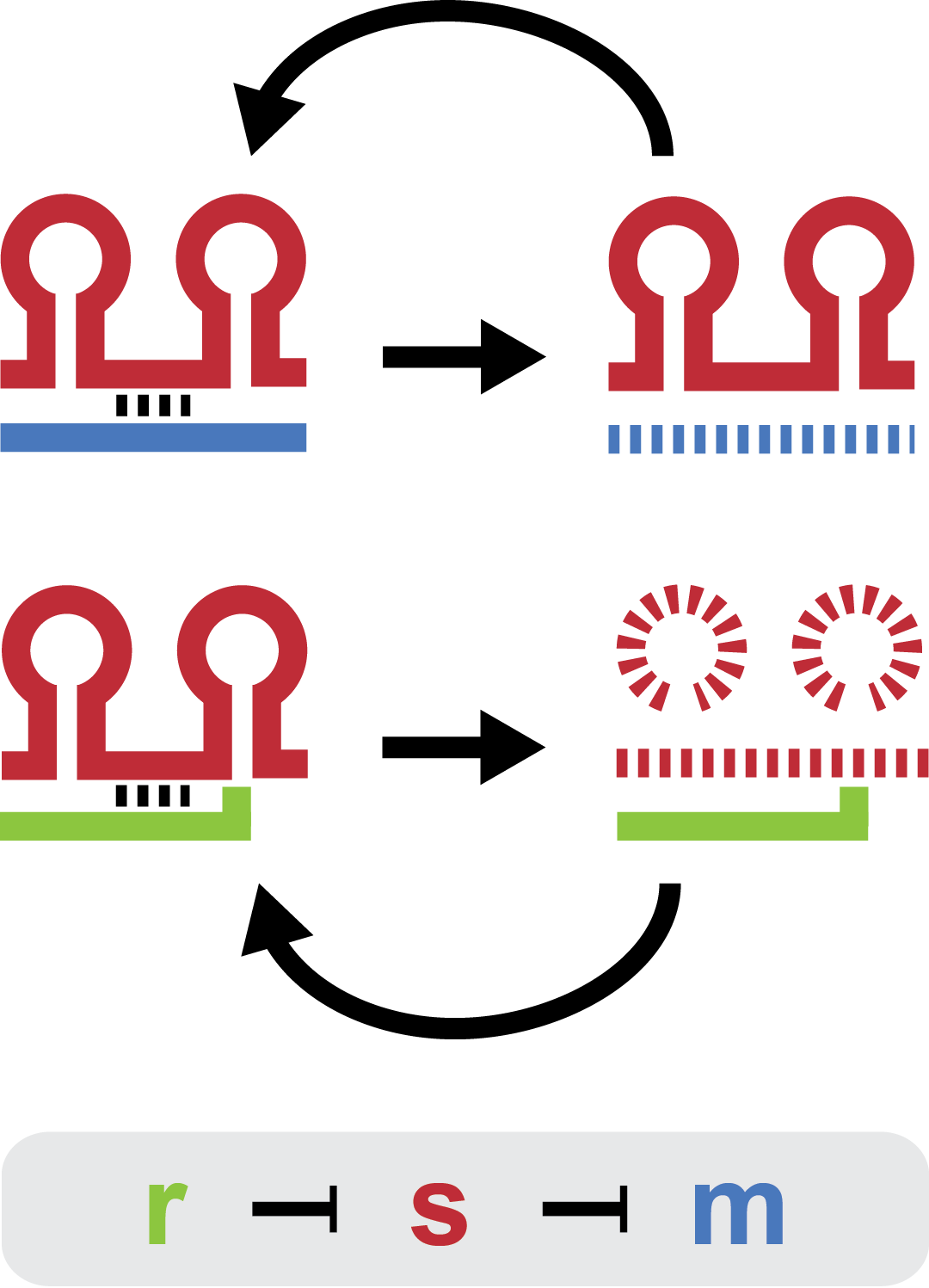
[[File:DTU2011_project_fig1.png|100px|frameless|right|Two-level sRNA regulation. Blue is any target mRNA, red is sRNA and green is trap-RNA.]] | [[File:DTU2011_project_fig1.png|100px|frameless|right|Two-level sRNA regulation. Blue is any target mRNA, red is sRNA and green is trap-RNA.]] | ||
| - | Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. Such applications include convenient gene silencing and fine-tuning of gene expression, which are currently cumbersome processes restricted to well studied bacteria. We have investigated a novel type of RNA regulation, where the inhibition caused by a small RNA is relieved by another small RNA called trap-RNA. We explore the possibility of using the system to uniquely target and repress any gene of interest, providing unprecedented specificity and control of gene silencing. | + | Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. Such applications include convenient gene silencing and fine-tuning of gene expression, which are currently cumbersome processes restricted to well studied bacteria. We have investigated a novel type of RNA regulation based on the [[https://2011.igem.org/Team:DTU-Denmark/Background_the_natural_system|chitobiose system]], where the inhibition caused by a small RNA is relieved by another small RNA called trap-RNA. We explore the possibility of using the system to uniquely target and repress any gene of interest, providing unprecedented specificity and control of gene silencing. |
<html> | <html> | ||
Revision as of 19:16, 21 September 2011
Project: Overview
Contents |
Abstract
Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. Such applications include convenient gene silencing and fine-tuning of gene expression, which are currently cumbersome processes restricted to well studied bacteria. We have investigated a novel type of RNA regulation based on the [system], where the inhibition caused by a small RNA is relieved by another small RNA called trap-RNA. We explore the possibility of using the system to uniquely target and repress any gene of interest, providing unprecedented specificity and control of gene silencing.
Bioinformatic
A bioinformatic study was performed to investigate the possibilities of engineering the trap-RNA system to target any gene. The study elucidates interesting features of sequence and secondary structure conservation guiding future application.
Experiments 1
Verifying that the envisioned small RNA based gene silencing is possible. Plasmids containing and strains deleted for the components were constructed providing a biological model.
Experiments 2
The dynamic range of the araBAD promoter was expanded.
Modeling
A framework for characterization was developed to guide rational design and test hypotheses. Steady state analysis revealed that each trap-RNA system has a characteristic fold repression.
Data
The data page provides a description of the constructed BioBricks and how they work.
 "
"