Team:ETH Zurich/Modeling/Analysis
From 2011.igem.org
(→Protein Degradation Rates) |
(→Stochastic Simulations) |
||
| Line 123: | Line 123: | ||
For the stochastic simulations we used the software Dizzy, to which we gave as an input file an SMBL file with the description of our system. Moving from deteministic to stochastic simulations, we had to change several things in the SBML file. For instance, we had to convert all the concentrations into number of molecules (and round them) and also separate the ODEs for the species into ODEs for distinct reaction channels (i.e. separate degradation from activation/repression) etc. | For the stochastic simulations we used the software Dizzy, to which we gave as an input file an SMBL file with the description of our system. Moving from deteministic to stochastic simulations, we had to change several things in the SBML file. For instance, we had to convert all the concentrations into number of molecules (and round them) and also separate the ODEs for the species into ODEs for distinct reaction channels (i.e. separate degradation from activation/repression) etc. | ||
| - | We started with a deterministic simulation to take the deterministic steady states of the species, after 5000 min(the acetaldehyde amount was the one that leads to the maximal GFP value, i.e the peak of the band). Starting from deterministic steady states, we performed a stochastic simulation with Gillespie-direct algorithm for 10000 min in order to get the stochastic steady states. After that, we restarted the stochastic simulation having the stochastic steady state values as our initial and simulated for 100 000 more minutes (storing the GFP values every | + | We started with a deterministic simulation to take the deterministic steady states of the species, after 5000 min(the acetaldehyde amount was the one that leads to the maximal GFP value, i.e the peak of the band). Starting from deterministic steady states, we performed a stochastic simulation with Gillespie-direct algorithm for 10000 min in order to get the stochastic steady states. After that, we restarted the stochastic simulation having the stochastic steady state values as our initial and simulated for 100 000 more minutes (storing the GFP values every minute). We collected all the GFP values from the last simulation and plotted them in a histogram (150 bins). |
| - | We see in Figure 1 that we obtain a Gaussian distribution of GFP number of molecules at the steady state, with a mean of | + | We see in Figure 1 that we obtain a Gaussian distribution of GFP number of molecules at the steady state, with a mean of 23491 molecules and standard deviation of 667 molecules. If we convert this value to concentration we get 19.5046 μM, which is very close to what we obtained deterministically. We see that the system keeps fluctuating -very closely to its steady state, with no large jumps away from it. The fact that the Gaussian distribution is unimodal tells us that our system is monostable. From this analysis we confirmed what we concluded before and that is the robustnes of our system, especially of the GFP band. We can now be sure that for a certain acetaldehyde concentration once the band appears, it won't start fading away. |
|} | |} | ||
Revision as of 13:11, 24 October 2011
| System Analysis |
| ||
| We wanted to analyse the effect of the parameters on the output of our system. We achieved this by looking at how the characteristics of the GFP band change when we explore the parameter space of a certain constant and at the sensitivity of GFP to the value of the toxic input substance (acetaldehyde or xylene) . | |||
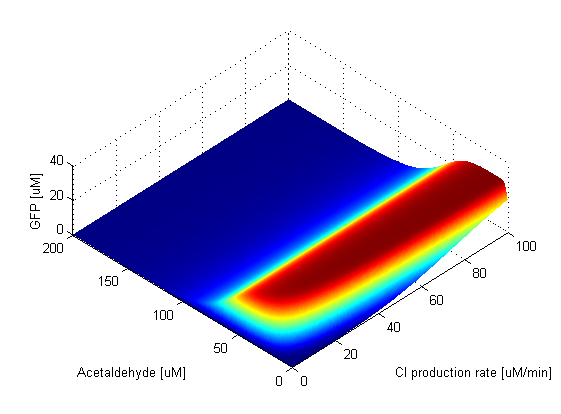
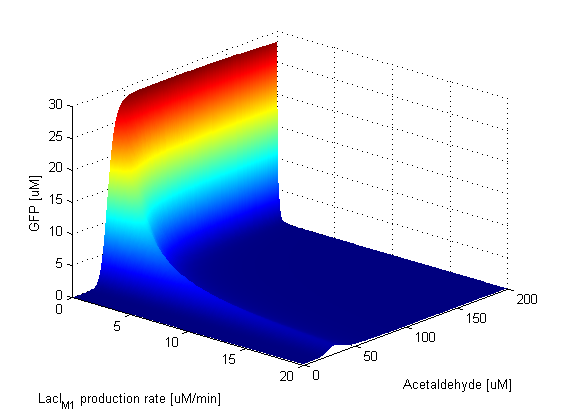
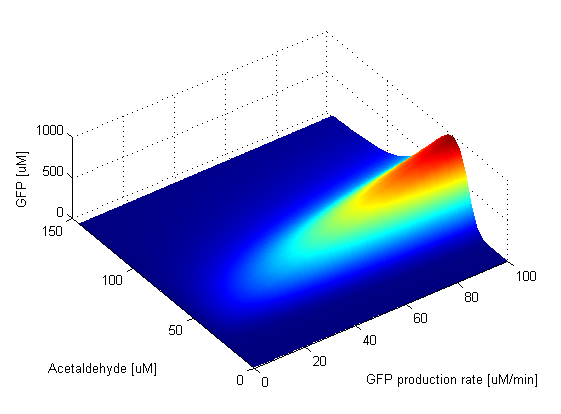
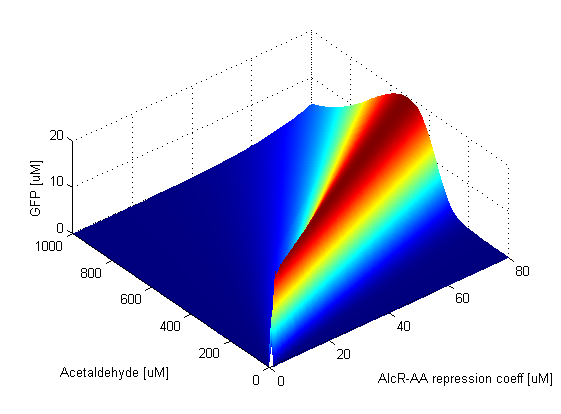
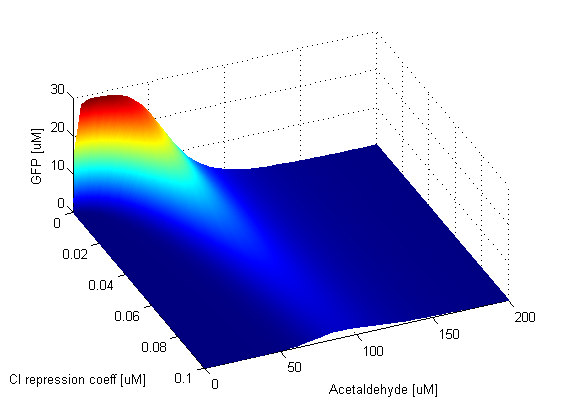
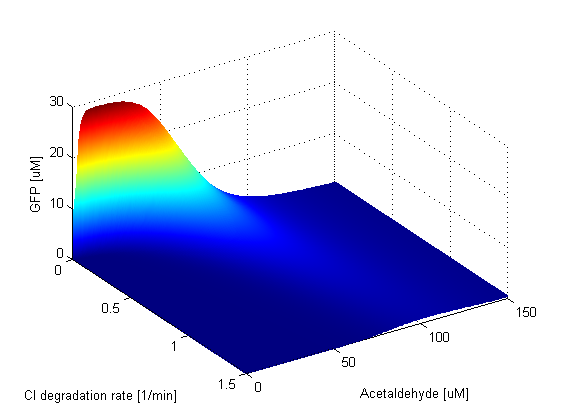
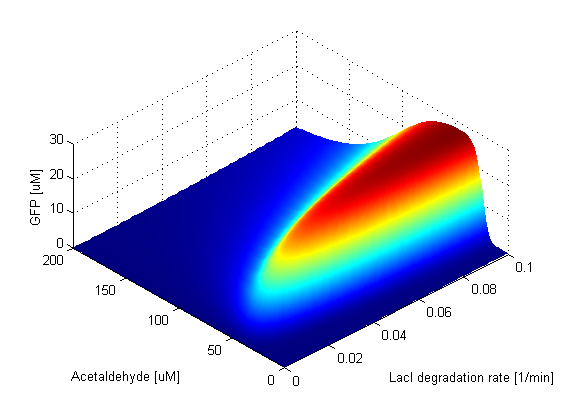
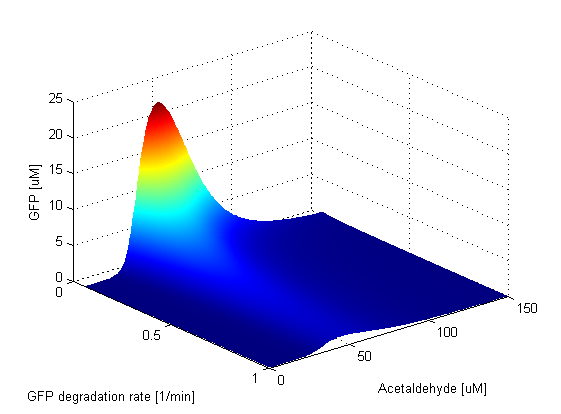
Parameter SweepsFor the parameters that belong to the band detector module, we explored their parameter spaces to quantify how they affect the features of the GFP band . As we varied each parameter (while keeping the rest constant), we varied also the acetaldehyde input and monitored the GFP output. We analyzed the parameter space only for the model that uses acetaldehyde as an input parameter. In both models the band is affected in the same way by the parameters of the band detector module. The sensor mechanism can only cause shifts in the band but the band detector module itself is unaffected by the input of the system. The parameter sweeps for the xylene model would therefore be similar to the acetaldehyde model, with differences only for the parameters involved in the sensor mechanism. In order to speed up the simulations, we used Maple to derive the steady-state formulas for all the relevant species. In this way we were able to obtain the steady-state GFP value simply by knowing the input concentration of Acetaldehyde, without the need to use an ODE solver. Thus we were able to reduce the simulation time from about 30 minutes to under one minute for one parameter sweep. The following figures show how the band changes with the variation of protein production rates, repression coefficients and degradation rates.
Protein Synthesis Rates
Repression Coefficients
Protein Degradation Rates
| ||||||||||||||||||||||||||||
Coupled parameter variationWith doing the parameter sweeps for individual parameters, we saw how robust our system is with respect to a particular paramter. But in reality, when parameters vary, they usually don't vary alone, but they are coupled (i.e more parameters vary simultaneously). One way to incorporate this fact in our model is to couple the expression rates of the proteins according to the plasmid they are expressed on and vary them together. |
Stochastic SimulationsBesides the parameter space search, we performed another type of analysis in order to verify that our system is robust and that it is not bistable. We were especially interested at the GFP band and whether it is always there. We did stochastic simulations to see how our system reacts to noise and how it responds to perturbations. For a certain acetaldehyde concentration (which gives the peak of the band) we wanted to see how the GFP peak value varies over long time period when we consider stochasticity. For the stochastic simulations we used the software Dizzy, to which we gave as an input file an SMBL file with the description of our system. Moving from deteministic to stochastic simulations, we had to change several things in the SBML file. For instance, we had to convert all the concentrations into number of molecules (and round them) and also separate the ODEs for the species into ODEs for distinct reaction channels (i.e. separate degradation from activation/repression) etc. We started with a deterministic simulation to take the deterministic steady states of the species, after 5000 min(the acetaldehyde amount was the one that leads to the maximal GFP value, i.e the peak of the band). Starting from deterministic steady states, we performed a stochastic simulation with Gillespie-direct algorithm for 10000 min in order to get the stochastic steady states. After that, we restarted the stochastic simulation having the stochastic steady state values as our initial and simulated for 100 000 more minutes (storing the GFP values every minute). We collected all the GFP values from the last simulation and plotted them in a histogram (150 bins).
|
Sensitivity AnalysisSensitivity analysis is a technique that studies the change of the output (or any observable) of a certain function with the variation of a certain parameter. It gives us an overview of how sensitive the model is with respect to the parameter, i.e. what the impact of the parameter is. The sensitivity is defined as the partial differential equation of the observable with respect to a certain parameter. We performed sensitivity analysis with respect to acetaldehyde or xylene. For different input concentrations we monitored the change in the GFP output (dGFP/dAcetaldehyde or dGFP/dXylene).
|
 "
"