Team:Harvard/Project
From 2011.igem.org
(→Clinical Applications of Zinc Fingers) |
|||
| Line 70: | Line 70: | ||
=Clinical Applications of Zinc Fingers= | =Clinical Applications of Zinc Fingers= | ||
| - | + | '''Colorblindness (Red Opsin)''' | |
| - | + | *Goal: Produce functional red opsin photoreceptor proteins in the eye | |
| - | + | *Method: Insertion of functional red opsin gene (''OPN1LW'' [http://genome.ucsc.edu/cgi-bin/hgGene?hgg_geneuc004fjz.3&hgg_protP04000&hgg_chromchrX&hgg_start153409724&hgg_end153424505&hgg_typeknownGene&dbhg19&hgsid206379197 <sup>1</sup>]) upstream of normal locus in patient lacking the gene | |
| - | + | '''Inherited High Cholesterol (Familial Hypercholesterolemia)''' | |
| - | + | *Goal: Produce functional LDLR protein to remove LDL cholesterol from the blood | |
| - | + | *Method: Insertion of functional ''LDLR'' gene upstream of nonfunctional allele | |
| - | + | '''Cancer (Myc Oncogene)''' | |
| - | + | *Goal: Knock out the oncogenic protein product and stop cancerous proliferation | |
| - | + | *Method: Targeted disruption (deletion) in mutated oncogene | |
=Technological Applications= | =Technological Applications= | ||
Revision as of 02:50, 18 October 2011
Overview | Design | Synthesize | Test | Zinc Finger Background | Protocols
Contents |
Our Project
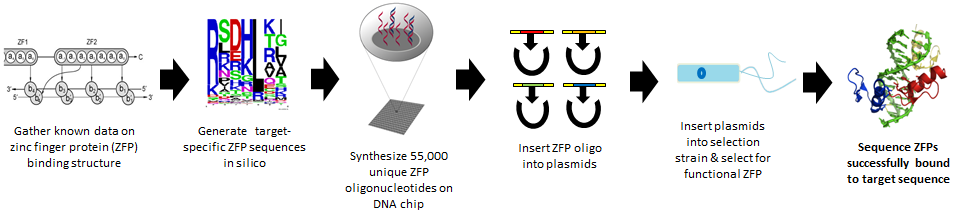
1. Design: use a bioinformatics approach to predict 55,000 zinc finger sequences
- Targeted against six DNA sequences for three diseases
2. Synthesize: use chip-based DNA synthesis to make all 55,000 sequences in one tube
3. Test: use a metabolic selection system to test which zinc finger sequences successfully bind DNA
Abstract
Gene therapy is a powerful approach for the treatment of disease; however, current therapies using viral vectors carry significant risk of tumorigenesis due to their use of non-specific gene insertion. To meet this challenge, we engineered zinc finger proteins (ZFPs), which are tailored to bind to DNA with high specificity, enabling precise genome editing. Our group has developed a foundational technology for synthesizing nearly 55,000 unique ZFPs using chip-based DNA synthesis based on bioinformatic analysis and for identifying the best binder using a novel genomically encoded 1-hybrid genetic selection scheme in a massively multiplexed fashion. Further, we employed multiplex automated genome engineering (MAGE) for facile editing of the E. coli genome enabling rapid modification of ZFP target sites, gene knockouts and silent codon substitutions. These tools allow for low-cost creation of ZFPs targeting any endogenous human gene, which will increase the accessibility of customized genome editing for gene therapy.
Zinc Finger Background
What are Zinc Finger Proteins (ZFPs)?
Function
ZFPs are found commonly in nature as a class of special transcription factors that bind to DNA, thus regulating gene expression. Zinc finger function was first studied using zinc finger protein Zif268.
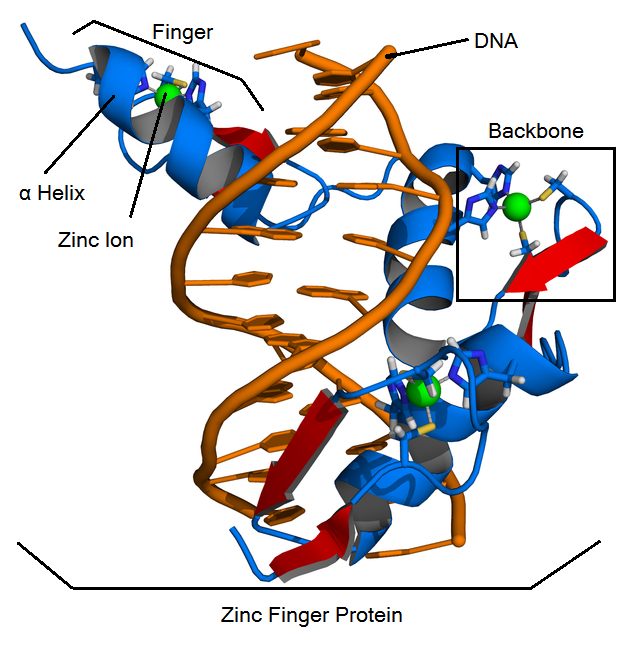
Structure
ZFPs consist of smaller subunits called "fingers" which each contain a zinc finger binding helix that binds to unique DNA sequences. These fingers are linear and linked together by the "zinc finger backbone", a series of approximately 21 amino acids.
- Cis2His2 ZFPs have three main structural components:
- Zinc finger binding helix
- Linker region
- Zinc ion that is coordinated by two cysteine residues and two histidine residues.
Helpful Zinc Finger Links
[http://en.wikipedia.org/wiki/Zinc_finger Zinc Fingers on Wikipedia]
- A more detailed introduction to zinc fingers.
[http://compbio.cs.princeton.edu/zf/ Predicting DNA Recognition by C2H2 Zinc Finger Proteins]
- A program useful for predicting how well a given amino acid sequence will bind to a given DNA sequence
[http://www.zincfingers.org/default2.htm The Zinc Finger Consortium]
- Information & helpful resources for zinc fingers
[http://www.jounglab.org/ Joung Lab]
- Information about Dr. Joung's extensive work with zinc fingers
Project Overview
Zinc fingers are specialized proteins that bind to DNA. Due to their ability to target highly specific DNA sequences, zinc fingers offer great potential for gene therapy and personalized medicine: recently, they were shown to be effective in conferring HIV resistance and treating hemophilia in mice. In the past, however, designing new zinc fingers--a necessity for individualized gene therapy--has been prohibitively expensive and time consuming.
For our iGEM project, we are creating and testing thousands of zinc fingers simultaneously at a cost feasible for most labs. To do so, we harnessed two novel synthetic biology technologies: chip-based synthesis, which allows up to millions of DNA strands to be synthesized concurrently, and MAGE (multiplex automated genome engineering), which makes possible direct edits of the genome of organisms, rather than using the small, cumbersome plasmids that are common in labs today.
As the structure and binding interactions of zinc fingers are not yet understood, our project also utilizes bioinformatics and computational analysis of the limited existing data to make “educated guesses” to generate zinc fingers that could bind to DNA for genes that cause colorblindness, some types of cancer, and high cholesterol.
Thus, our zinc fingers and their clinical applications are a new technology, and while we hope that our new zinc fingers work, the more important goal is maximizing efficiency and decreasing cost while utilizing new technology: we anticipate that future iGEM teams will find great use for chip-based synthesis and MAGE.
Clinical Applications of Zinc Fingers
Colorblindness (Red Opsin)
- Goal: Produce functional red opsin photoreceptor proteins in the eye
- Method: Insertion of functional red opsin gene (OPN1LW [http://genome.ucsc.edu/cgi-bin/hgGene?hgg_geneuc004fjz.3&hgg_protP04000&hgg_chromchrX&hgg_start153409724&hgg_end153424505&hgg_typeknownGene&dbhg19&hgsid206379197 1]) upstream of normal locus in patient lacking the gene
Inherited High Cholesterol (Familial Hypercholesterolemia)
- Goal: Produce functional LDLR protein to remove LDL cholesterol from the blood
- Method: Insertion of functional LDLR gene upstream of nonfunctional allele
Cancer (Myc Oncogene)
- Goal: Knock out the oncogenic protein product and stop cancerous proliferation
- Method: Targeted disruption (deletion) in mutated oncogene
Technological Applications
- The novel methods we employed in our project have the potential to revolutionize synthetic biology practices, and the way that future iGEM competitions are conducted. To learn more about the technological applications of our project, please see our Technology page.
Detailed Project Description
Gene therapy is a powerful approach for the treatment of disease, and holds great potential for success. While gene therapy has progressed significantly since its conception, much of its projected potential remains untapped, while existing therapeutic advancements are still fraught with serious problems. Many of these roadblocks to existing therapies are caused by non-specific gene insertion (as through viral vectors), which hinders therapeutic viability and can lead to unintended side effects including cancer. In recent attempts to overcome this obstacle, however, one particular approach has shown much promise. This approach utilizes engineered zinc finger proteins, which have been shown to edit the genome with dramatically increased accuracy (Perez et al 2008, Li et al 2011). Zinc finger proteins are naturally-occurring protein domains composed of multiple individual zinc finger subdomains. Although the individual subdomains (“fingers”) occur in a variety of structural motifs, the classic Cys2His2 (C2H2) motif is among the best characterized and can be observed in a variety of transcription factors. The C2H2 motif (see Fig. 1) consists of a beta-sheet conjugated to an alpha helix, and it is structurally coordinated through interactions between two cysteine and two histidine residues with a single zinc ion. Many eukaryotic transcription factors utilize C2H2 zinc finger domains, which possess a unique capacity to bind to specific DNA sequences. Zinc finger proteins have received increasing attention in recent research for their DNA-binding ability and specificity, which offers a solution to non-specificity in gene therapy and could thus bring the scientific community a step closer to realizing many novel therapeutic applications. Recent studies, for instance have demonstrated that DNA-binding zinc fingers coupled with Type IIs restriction endonucleases such as Fok1 can be harnessed as effective tools to genetically treat hemophilia and confer HIV resistance in mouse models (Perez et al 2008, Li et al 2011).While many different natural zinc finger proteins can be employed for synthetic biology-based tasks, engineered zinc finger proteins are most commonly composed of an array of three zinc fingers based on Zif268. Zif268 is a well-characterized mammalian early response transcription factor also known as EGR-1 that was discovered in mice. Zif268 has three C2H2 finger subunits, and each finger binds a specific 3-nucleotide base pair (bp) triplet on a DNA strand. Composed of an array of three fingers, Zif268 binds to three consecutive DNA triplets that in total comprise a 9-bp binding site. This 3-finger domain, 9-bp specificity is a commonly occurring theme in zinc finger literature, although it is by no means a rule. Natural and engineered zinc finger arrays may contain fewer or more zinc fingers and bind DNA sequences of corresponding length and specificity. While any given zinc finger might bind a certain 3-bp DNA sequence with exquisite specificity, even the most specific 3-bp binding event is of little practical value for gene therapy, as a given 3-bp sequence may appear countless times within a given genome. Thus, for practical use in drug delivery or gene therapy, zinc fingers must be engineered to bind more specifically to DNA sequences. Such increased specificity can be achieved through the creation of multi-finger arrays that bind to specific 9-bp sequences. With this in mind, research has focused on creating modular zinc finger subunits, which can be combined into multi-finger arrays. Three-finger arrays, for instance, are tailored to bind to a specific 9-bp sequence through the selection of appropriate zinc fingers to bind to each DNA triplet.
Different studies have suggested different models of binding and varying levels of success for zinc finger modularity. Thus, through an approach utilizing data-mining, binding models, and comprehensive bioinformatics, in conjunction with next-generation oligo synthesis technology, we hope to develop a method for engineering zinc finger proteins that can bind to any arbitrarily-selected DNA sequence. The success of such an engineering method would, in turn, facilitate clinical advancements by promoting highly-specific, targeted gene therapies, and would promote personalized medicine by allowing the production of zinc finger-based therapies tailored to unique genomic sequences.
References
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, Murphy SL, Finn JD, Khazi FR, Zhou S, Paschon DE, Rebar EJ, Bushman FD, Gregory PD, Holmes MC, High KA (2011). In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature, doi: 10.1038/nature10177 [Epub ahead of print].
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH (2008). Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nature Biotechnology, 26(7), 808-16.
 "
"