Team:Washington/Alkanes/Results
From 2011.igem.org
(→GCMS confirms The PetroBrick enables diesel production from sugar using E. coli) |
(→GCMS confirms The PetroBrick enables diesel production from sugar using E. coli) |
||
| Line 6: | Line 6: | ||
='''GCMS confirms The PetroBrick enables diesel production from sugar using ''E. coli'''''= | ='''GCMS confirms The PetroBrick enables diesel production from sugar using ''E. coli'''''= | ||
| - | We | + | We performed GC-MS analysis on extracts from cell cultures expressing both [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590032 Acyl-ACP Reductase] (AAR) and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590031 Aldehyde Decarbonylase] (ADC) (this composite part is designated [http://partsregistry.org/Part:BBa_K590025 The PetroBrick]). To act as controls and in order to show that the alkane production system is working as expected, we also analyzed cell cultures expressing either only AAR, or only ADC. |
Revision as of 01:06, 29 September 2011
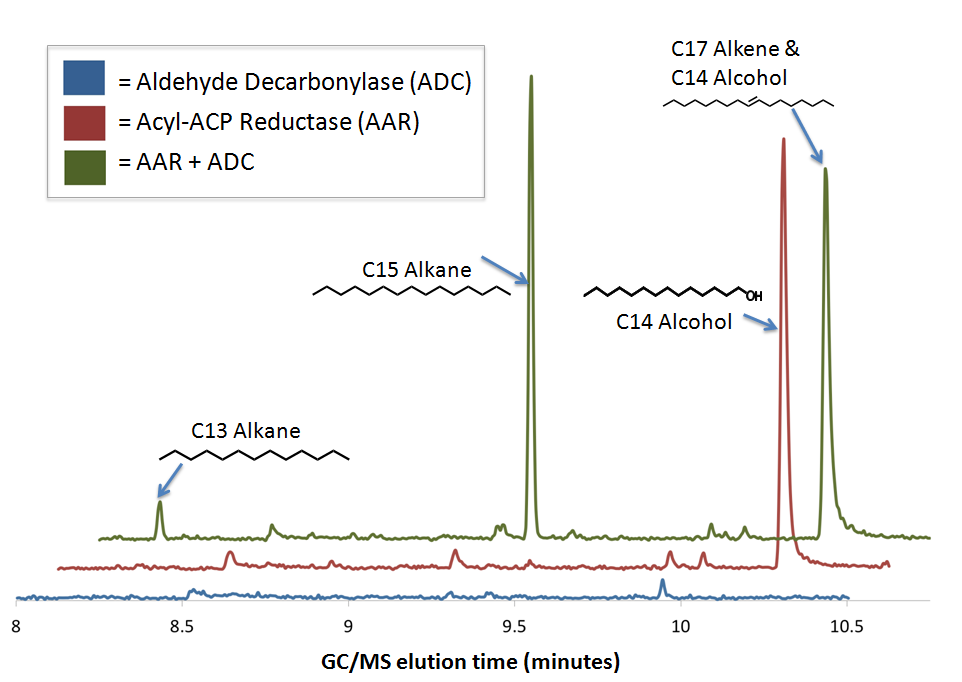
GCMS confirms The PetroBrick enables diesel production from sugar using E. coli
We performed GC-MS analysis on extracts from cell cultures expressing both [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590032 Acyl-ACP Reductase] (AAR) and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590031 Aldehyde Decarbonylase] (ADC) (this composite part is designated [http://partsregistry.org/Part:BBa_K590025 The PetroBrick]). To act as controls and in order to show that the alkane production system is working as expected, we also analyzed cell cultures expressing either only AAR, or only ADC.
No significant hydrocarbons peaks in samples extracted from cells expressing only ADC. The blue GCMS chromatagram trace on the bottom show no significant peaks are found on the ADC only control within the time range that we expect alkane signal based upon GC-MS runs on chemical standards (8-10.5 minutes). This is expected, as E. coli does not normally produce any long chain length aldehyde substrates.
C16 alcohols are observed when AAR is expressed on its own. The red GCMS chromatagram trace in the middle has a significant peak at 10.2 minutes corresponding to the C16 alcohol (as confirmed by comparison of the peak's MS spectra to a reference library) is observed when AAR is expressed in MG1655 E. coli. The production of C16 alcohols in cells expressing AAR (but not in cells expressing only ADC) is consistent with AAR reducing even chain length Acyl-ACPs into even chain length fatty aldehydes, which are further reduced by aldehyde dehydrogenases to the alcohol.
The PetroBrick Works! C13, C15, and C17 alkanes are produced when both AAR and ADC are expressed.
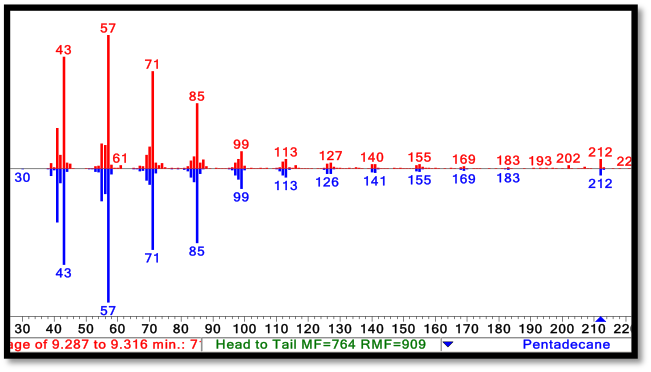
The green GCMS chromatagram trace in the middle has a significant new peaks corresponding to the C13 (8.2min) and C15 (9.2min) alkanes, as well as a peak at 10.2 minutes that contains both the C14 alcohol and the C17 alkene. The fact that the C17 alkene peak overlaps with the C14 alcohol peak makes exact quantification of C17 akene yields impossible, but we have sufficient evidence to determine that both molecules are present in the 10.2 minute peak( the earlier section of the peak has an MS spectra consistent with the C16 alcohol, and the later section is consistent with the C17 alkene). The C13 and C15 alkane peaks showed MS spectra highly consistent with C13 and C15 alkane reference spectra(for these spectra, as well as the C17 alkene spectrum, refer to our Spectra Page.). In addition, both alkane molecules eluted off of the GC at the same time as C13 and C15 alkane standards, further increasing confidence that the 8.2 min and 9.2 min peaks do correspond to the C13 and C15 alkanes. Odd chain length alkane production is what is expected from our system, as ADC removes a carbonyl group from even chain length aldehydes produced by AAR, yielding an odd chain length alkane. Using the PetroBrick, we can turn simple sugars into diesel, a fuel fully compatible with modern infrastructure.
Initial Quantization of Alkane Production
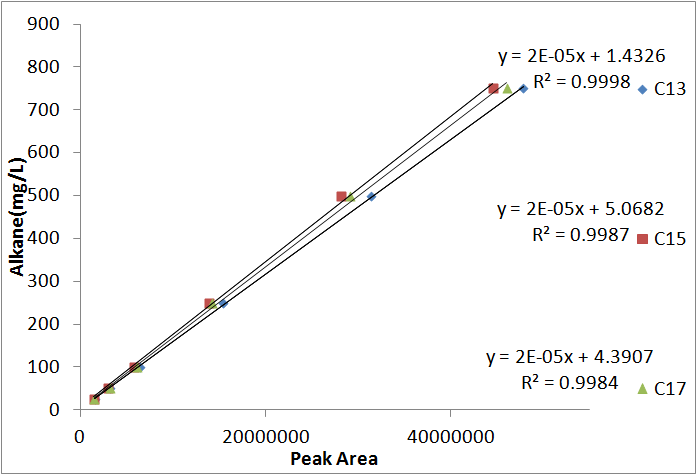
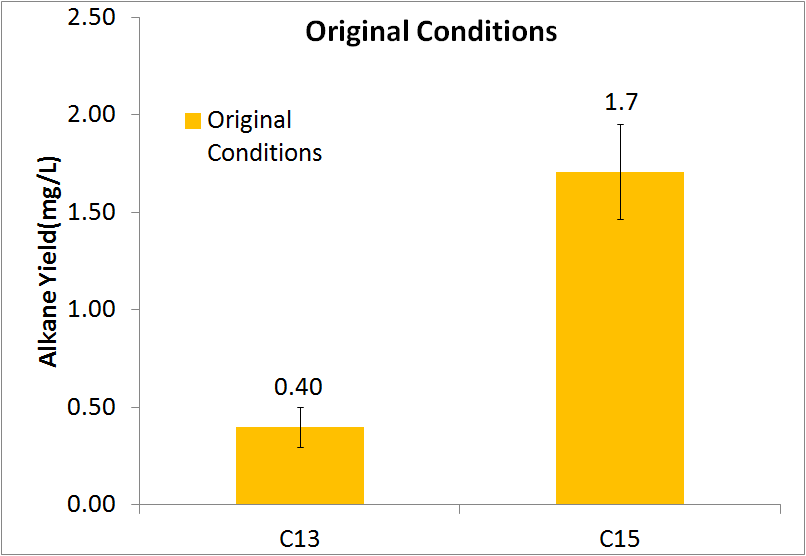
In order to be able to know how much alkane was being produced by our E. coli, we spiked known amounts of alkane into cell cultures known to not produce alkanes. We then extracted using ethyl acetate, and analyzed extracts using GC-MS. Since peak area corresponds to the amount of each substance present, we used these GC plots to make a standard curve that allows us to convert peak area into an absolute yield. To determine how much alkane was being produced by the Petrobrick, we grew up 3 MG1655 cell cultures transformed with the PetroBrick in M9 production media (link), and analyzed using GC-MS.
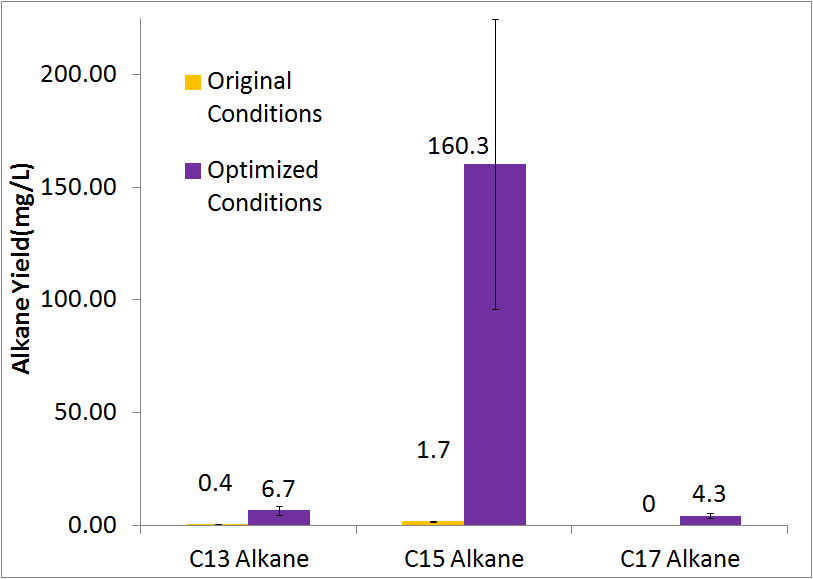
Optimized Production
By optimizing the method of growth, media, vectors, and cell line we were able to successfully increase alkane yield over 80-fold. Note that this improvement doesn't include C17 alkene production, as we were unable to quantify the C17 alkene peak due to co-elution with the C14 alcohol. The parameters we adjusted for optimization are discussed in detail at our systems optimization page under future directions, as this is an on going process.
 "
"