|
|
| Line 20: |
Line 20: |
| | <li class="toclevel-2"><a href="#note"><span class="tocnumber">2.1</span> <span class="toctext">Notes on promoter characterization</span></a></li> | | <li class="toclevel-2"><a href="#note"><span class="tocnumber">2.1</span> <span class="toctext">Notes on promoter characterization</span></a></li> |
| | <li class="toclevel-2"><a href="#rbs"><span class="tocnumber">2.2</span> <span class="toctext">RBSs from the community collection</span></a></li> | | <li class="toclevel-2"><a href="#rbs"><span class="tocnumber">2.2</span> <span class="toctext">RBSs from the community collection</span></a></li> |
| - | <li class="toclevel-2"><a href="#pLux"><span class="tocnumber">2.3</span> <span class="toctext">pLux - a 3OC6-HSL-in PoPs-out device</span></a></li> | + | <li class="toclevel-2"><a href="#pLux"><span class="tocnumber">2.3</span> <span class="toctext">p<sub>Lux</sub> - a 3OC<sub>6</sub>-HSL-in PoPs-out device</span></a></li> |
| - | <li class="toclevel-2"><a href="#pTet"><span class="tocnumber">2.4</span> <span class="toctext">pTet - BBa_R0040</span></a></li> | + | <li class="toclevel-2"><a href="#pTet"><span class="tocnumber">2.4</span> <span class="toctext">p<sub>Tet</sub> - BBa_R0040</span></a></li> |
| | <li class="toclevel-2"><a href="#LuxI"><span class="tocnumber">2.5</span> <span class="toctext">LuxI - BBa_C0061</span></a></li> | | <li class="toclevel-2"><a href="#LuxI"><span class="tocnumber">2.5</span> <span class="toctext">LuxI - BBa_C0061</span></a></li> |
| | <li class="toclevel-2"><a href="#AiiA"><span class="tocnumber">2.6</span> <span class="toctext">AiiA - BBa_C0060</span></a></li> | | <li class="toclevel-2"><a href="#AiiA"><span class="tocnumber">2.6</span> <span class="toctext">AiiA - BBa_C0060</span></a></li> |
| Line 62: |
Line 62: |
| | <p>For this reason, RPUs might not be reliable when comparing the same promoter with different RBSs because of the un-modularity of the RBS. In order to asses what's the effect of RBS 'un-modularity' on RPUs reliability, we have built a set of four constitutive promoters (BBa_J23101) followed by one of the four RBSs tested. These parts were used to evaluate RBS efficiency. Data were collected and analyzed as described in 'Measurements' and 'Data analysis' sections. RPUs and Synthesis rate per cell [AUr] were computed and results are summarized in the table below.</p> | | <p>For this reason, RPUs might not be reliable when comparing the same promoter with different RBSs because of the un-modularity of the RBS. In order to asses what's the effect of RBS 'un-modularity' on RPUs reliability, we have built a set of four constitutive promoters (BBa_J23101) followed by one of the four RBSs tested. These parts were used to evaluate RBS efficiency. Data were collected and analyzed as described in 'Measurements' and 'Data analysis' sections. RPUs and Synthesis rate per cell [AUr] were computed and results are summarized in the table below.</p> |
| | | | |
| - | <p><br><em><b>NB</b>: in the RPU computation, the J23101-RBS34-mRFP-TT construct has been considered as the reference standard. With this assumption, RPUs are identical to the estimated RBS efficiency.</em></p></div> | + | <p><br><em><b>NB</b>: in the RPU computation, the J23101-RBS34-mRFP-TT (<A HREF="http://partsregistry.org/wiki/index.php/Part:BBa_J23101">BBa_J23101</a>) in low copy plasmid <A HREF="http://partsregistry.org/wiki/index.php/Part:pSB4C5">pSB4C5</a> construct has been considered as the reference standard. With this assumption, RPUs are identical to the estimated RBS efficiency.</em></p></div> |
| | | | |
| | <table align='center'><tr><td width='50%' > | | <table align='center'><tr><td width='50%' > |
| Line 119: |
Line 119: |
| | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516214 "> BBa_K516214 </a> (wiki name: E16 ) pTet-RBS34-LuxI </li></ul></ol> | | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516214 "> BBa_K516214 </a> (wiki name: E16 ) pTet-RBS34-LuxI </li></ul></ol> |
| | | | |
| | + | <br><br> |
| | + | <p align='justify'><em>Though these parts don't have a transcriptional terminator, they have been characterized in low copy plasmid <A HREF="http://partsregistry.org/wiki/index.php/Part:pSB4C5">pSB4C5</a>, that contains the BBa_B0054 terminator. This choice is motivated by the need to reproduce the exact experimental context of the final circuit, as described in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Solution#Circuit_design'>solution section</a>. |
| | + | </em></p> |
| | + | <br><br> |
| | + | <p align='justify'> |
| | + | LuxI has been characterized in terms of enzymatic activity under the regulation of p<sub>Tet</sub> promoter. |
| | + | </p> |
| | <p align='justify'> | | <p align='justify'> |
| - | LuxI has been characterized in terms of enzymatic activity under the regulation of pet promoter. K<sub>M,LuxI</sub> and V<sub>max</sub> parameters representing its activity have been estimated and the promoter strength (represented by a synthetic parameter α<sub>pTet</sub> for every pTet-RBS combination) at full induction (100 ng/ml) has been estimated too with a simultaneous fitting of the available data.
| + | K<sub>M,LuxI</sub> and V<sub>max</sub> parameters representing its activity have been estimated and the promoter strength (represented by a synthetic parameter α<sub>pTet</sub> for every pTet-RBS combination) at full induction (100 ng/ml) has been estimated too with a simultaneous fitting of the available data. |
| | | | |
| | </p> | | </p> |
| Line 130: |
Line 137: |
| | | | |
| | LuxI has been characterized through the Biosensor BBa_T9002 (see <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#introduction_to_T9002'>modelling section</a>). | | LuxI has been characterized through the Biosensor BBa_T9002 (see <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#introduction_to_T9002'>modelling section</a>). |
| - | <br>The HSL synthesis rate has been evaluated according to the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Equations_for_gene_networks'>model equations</a>, properly adjusted. In particular, the ODE system is reported here: </p> | + | <br>The HSL synthesis rate has been evaluated according to the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Equations_for_gene_networks'>model equations</a>, properly adjusted. In particular, the ODE system describing the behavior of this measurement circuit is reported here: </p> |
| | | | |
| | <div style='text-align:justify'><div class="thumbinner" style="width: 850px;"> | | <div style='text-align:justify'><div class="thumbinner" style="width: 850px;"> |
| Line 139: |
Line 146: |
| | | | |
| | <p align='justify'> | | <p align='justify'> |
| - | Since the measurement systems are only assayed in the exponential growth phase, in the equation (3) N<<N<sub>max</sub> is assumed. | + | Since the measurement systems are only assayed in the exponential growth phase, in the equation (3) N<N<sub>max</sub> is assumed. |
| | The parameters N<sub>max</sub>, μ and γ<sub>HSL</sub> (see the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Results'>Results section</a> for more details) are known and summarized in the table below:</p> | | The parameters N<sub>max</sub>, μ and γ<sub>HSL</sub> (see the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Results'>Results section</a> for more details) are known and summarized in the table below:</p> |
| | | | |
| Line 207: |
Line 214: |
| | <!-- ----------------- --> | | <!-- ----------------- --> |
| | | | |
| - | <a name='pTetAiiA'></a><h2>AiiA expression cassette driven by aTc-inducible pTet promoter</h2> | + | <a name='pTetAiiA'></a><h2>AiiA expression cassette driven by aTc-inducible p<sub>Tet</sub> promoter</h2> |
| | <ol> | | <ol> |
| | <ul> | | <ul> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516220 "> BBa_K516220 </a> (wiki name: E24 ) pTet-RBS30-AiiA-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516220 "> BBa_K516220 </a> (wiki name: E24 ) p<sub>Tet</sub>-RBS30-AiiA-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516221 "> BBa_K516221 </a> (wiki name: E25 ) pTet-RBS31-AiiA-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516221 "> BBa_K516221 </a> (wiki name: E25 ) p<sub>Tet</sub>-RBS31-AiiA-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516222 "> BBa_K516222 </a> (wiki name: E26 ) pTet-RBS32-AiiA-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516222 "> BBa_K516222 </a> (wiki name: E26 ) p<sub>Tet</sub>-RBS32-AiiA-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516224 "> BBa_K516224 </a> (wiki name: E27 ) pTet-RBS34-AiiA-TT </li></ul></ol> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516224 "> BBa_K516224 </a> (wiki name: E27 ) p<sub>Tet</sub>-RBS34-AiiA-TT </li></ul></ol> |
| | | | |
| | <div align="right"><small><a href="#indice" title="">^top</a></small></div> | | <div align="right"><small><a href="#indice" title="">^top</a></small></div> |
| Line 261: |
Line 268: |
| | <a name='rbs'></a><h2>RBSs</h2> | | <a name='rbs'></a><h2>RBSs</h2> |
| | <p align='justify'> | | <p align='justify'> |
| - | <p><div align="justify">The complex RBS-promoter acts as a whole regulatory element and determines the amount of translated protein. RBSs have been reported to have an un-modular behavior, since the translational efficiency is not independent on the coding sequences, but variates as an effect of different mRNA structure stability [Salis et al., Nat Biotec, 2009]. In addition, it is not possible to separate the effects of the sole promoter and of the sole RBS on the total amount of mRFP produced. | + | <p><div align="justify">The complex RBS-promoter acts as a whole regulatory element and determines the amount of translated protein. RBSs have been reported to have an un-modular behavior, since the translational efficiency is not independent on the coding sequences, but variates as an effect of different mRNA structure stability [Salis et al., Nat Biotec, 2009]. In addition, it is not possible to separate the effects of the sole promoter and of the sole RBS on the total amount of protein produced. |
| | For this reason, every combination 'Promoter+RBS' was studied as a different regulatory element.</p> | | For this reason, every combination 'Promoter+RBS' was studied as a different regulatory element.</p> |
| | <p>The evaluation of RBS efficiency can be performed in a very intuitive fashion:</p> | | <p>The evaluation of RBS efficiency can be performed in a very intuitive fashion:</p> |
| Line 278: |
Line 285: |
| | | | |
| | | | |
| - | <li>measure the output of the circuits and calculate the RBS efficiency as the ratio of the output relative to the output of the circuit with the standard RBS. </li></div></ol><br> | + | <li>measure the output of the circuits and calculate the RBS efficiency as the ratio of the output relative to the output of the circuit with the standard RBS (<a href='http://partsregistry.org/wiki/index.php/Part: BBa_B0034'>BBa_B0034</a>). </li></div></ol><br> |
| | | | |
| | | | |
| - | <div align="justify"><p>This simple measurement system allows the quantification of RBS efficiency depending on the whole measurement system (i.e.: promoter and encoded gene). Today it has not still been completely validated the hypothesis that every functional module in a genetic circuit maintains its behavior when assembled in complex circuits, even if many researchers implicitly accept this hypothesis when performing characterization experiments.</p> | + | <div align="justify"><p>This simple measurement system allows the quantification of RBS efficiency depending on the experimental context (i.e.: promoter and encoded gene). Today it has not still been completely validated the hypothesis that every functional module in a genetic circuit maintains its behavior when assembled in complex circuits, even if many researchers implicitly accept this hypothesis when performing characterization experiments.</p> |
| | <p>To rationally assess the impact that this hypothesis has on the genetic circuit design and fine tuning, several measurement systems were built to evaluate the dependance of RBS modularity from the promoter or the coding sequence separately.</p> | | <p>To rationally assess the impact that this hypothesis has on the genetic circuit design and fine tuning, several measurement systems were built to evaluate the dependance of RBS modularity from the promoter or the coding sequence separately.</p> |
| - | <p>In particular, in order to investigate if RBS efficiency depends on the promoter, the same coding devices (RBSx-RFP-TT) were assembled downstream of different promoters (J23101, pTet, pLux). Measuring the system output and evaluating the RBS efficiency. The results are summarized in the table below:</div></p> | + | <p>In particular, in order to investigate if RBS efficiency depends on the promoter, the same coding devices (RBSx-RFP-TT) were assembled downstream of different promoters (J23101, p<sub>Tet</sub>, p<sub>Lux</sub>). The system output was measured and the RBS efficiency evaluated. The results are summarized in the table below:</div></p> |
| | <br> | | <br> |
| | <div align="center"><table class='data' width='70%'><tr> | | <div align="center"><table class='data' width='70%'><tr> |
| | <td class='row'><b>RBS</b></td> | | <td class='row'><b>RBS</b></td> |
| - | <td class='row'><b>eff<sub>pLux</sub></b><sup>*</sup></td> | + | <td class='row'><b>eff<sub>p<sub>Lux</sub></sub></b><sup>*</sup></td> |
| - | <td class='row'><b>eff<sub>pTet</sub></b><sup>*</sup></td> | + | <td class='row'><b>eff<sub>p<sub>Tet</sub></sub></b><sup>*</sup></td> |
| | <td class='row'><b>eff<sub>J23101</sub></b><sup>**</sup></td> | | <td class='row'><b>eff<sub>J23101</sub></b><sup>**</sup></td> |
| | <td class='row'><b>Declared efficiency</b></td> | | <td class='row'><b>Declared efficiency</b></td> |
| Line 302: |
Line 309: |
| | | | |
| | <br> | | <br> |
| - | <div align="justify">On the other end, to investigate the dependance of RBS modularity on the coding sequence, the same regulatory elements (pTet-RBSx) were assembled upstream of different encoded gene (mRFP, AiiA and LuxI). RBS efficiency was assessed and the results are summarized in the table below:</div><br> | + | <div align="justify">On the other hand, to investigate the dependance of RBS modularity on the coding sequence, the same regulatory elements (p<sub>Tet</sub>-RBSx) were assembled upstream of different encoded gene (mRFP, AiiA and LuxI). RBS efficiency was assessed and the results are summarized in the table below:</div><br> |
| | | | |
| | <div align="center"><table class='data' width='70%'><tr> | | <div align="center"><table class='data' width='70%'><tr> |
| Line 323: |
Line 330: |
| | <div align="justify"><p><sup>*</sup> The RBS efficiency for inducible devices expressing mRFP was estimated as the ratio of the AUCs (Area under the curve) of the induction curve of the system with the studied RBS and the B0034 reference: AUC<sub>P, RBSx</sub>/AUC<sub>P, B0034</sub></p> | | <div align="justify"><p><sup>*</sup> The RBS efficiency for inducible devices expressing mRFP was estimated as the ratio of the AUCs (Area under the curve) of the induction curve of the system with the studied RBS and the B0034 reference: AUC<sub>P, RBSx</sub>/AUC<sub>P, B0034</sub></p> |
| | <p><sup>**</sup> The RBS efficiency for constitutive promoters expressing mRFP was computed as the ratio between S<sub>cell<sub>P, RBSx</sub></sub>/S<sub>cell<sub>P, B0034</sub></sub></p> | | <p><sup>**</sup> The RBS efficiency for constitutive promoters expressing mRFP was computed as the ratio between S<sub>cell<sub>P, RBSx</sub></sub>/S<sub>cell<sub>P, B0034</sub></sub></p> |
| - | <p><sup>***</sup> The RBS efficiency for pTet promoter driving the expression of AiiA enzyme was computed as the ratio α<sub>pTet, RBSx</sub>/α<sub>pTet, B0034</sub> estimated for the measurement system pTet-RBSx-AiiA-TT. α<sub>pTet</sub> was estimated as described <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#AiiA'>here</a>. pTet was tested at full induction (100 ng/ml).</p> | + | <p><sup>***</sup> The RBS efficiency for p<sub>Tet</sub> promoter driving the expression of AiiA enzyme was computed as the ratio α<sub>p<sub>Tet</sub>, RBSx</sub>/α<sub>p<sub>Tet</sub>, B0034</sub> estimated for the measurement system p<sub>Tet</sub>-RBSx-AiiA-TT. α<sub>p<sub>Tet</sub></sub> was estimated as described <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#AiiA'>here</a>. p<sub>Tet</sub> was tested at full induction (100 ng/ml).</p> |
| - | <p><sup>****</sup> The RBS efficiency for promoters driving the expression of LuxI was computed as the ratio α<sub>pTet, RBSx</sub>/α<sub>pTet, B0034</sub> estimated from the measurement systems pTet-RBSx-LuxI. α<sub>pTet</sub> was estimated as described <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#LuxI'>here</a>. pTet was tested at full induction (100 ng/ml).</p></div> | + | <p><sup>****</sup> The RBS efficiency for promoters driving the expression of LuxI was computed as the ratio α<sub>p<sub>Tet</sub>, RBSx</sub>/α<sub>p<sub>Tet</sub>, B0034</sub> estimated from the measurement systems p<sub>Tet</sub>-RBSx-LuxI. α<sub>p<sub>Tet</sub></sub> was estimated as described <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#LuxI'>here</a>. p<sub>Tet</sub> was tested at full induction (100 ng/ml).</p></div> |
| | | | |
| | <br> | | <br> |
| Line 339: |
Line 346: |
| | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_J23101 "> BBa_J23101 </a> J101-RBS34-mRFP-TT </li> | | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_J23101 "> BBa_J23101 </a> J101-RBS34-mRFP-TT </li> |
| | </ul> | | </ul> |
| - | <li>pTet</li> | + | <li>p<sub>Tet</sub></li> |
| | <ul class="disc"> | | <ul class="disc"> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516230 "> BBa_K516230 </a> pTet-RBS30-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516230 "> BBa_K516230 </a> p<sub>Tet</sub>-RBS30-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516231 "> BBa_K516231 </a> pTet-RBS31-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516231 "> BBa_K516231 </a> p<sub>Tet</sub>-RBS31-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516232 "> BBa_K516232 </a> pTet-RBS32-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516232 "> BBa_K516232 </a> p<sub>Tet</sub>-RBS32-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_I13521 "> BBa_I13521 </a> pTet-RBS34-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_I13521 "> BBa_I13521 </a> p<sub>Tet</sub>-RBS34-mRFP-TT </li> |
| | </ul> | | </ul> |
| - | <li>pLux</li> | + | <li>p<sub>Lux</sub></li> |
| | <ul class="disc"> | | <ul class="disc"> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516330 "> BBa_K516330 </a> pLambda-RBS30-LuxR-T-pLux-RBS30-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516330 "> BBa_K516330 </a> pLambda-RBS30-LuxR-T-p<sub>Lux</sub>-RBS30-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516331 "> BBa_K516331 </a> pLambda-RBS30-LuxR-T-pLux-RBS31-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516331 "> BBa_K516331 </a> pLambda-RBS30-LuxR-T-p<sub>Lux</sub>-RBS31-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516332 "> BBa_K516332 </a> pLambda-RBS30-LuxR-T-pLux-RBS32-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516332 "> BBa_K516332 </a> pLambda-RBS30-LuxR-T-p<sub>Lux</sub>-RBS32-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516334 "> BBa_K516334 </a> pLambda-RBS30-LuxR-T-pLux-RBS34-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516334 "> BBa_K516334 </a> pLambda-RBS30-LuxR-T-p<sub>Lux</sub>-RBS34-mRFP-TT </li> |
| | </ul> | | </ul> |
| | </ul> | | </ul> |
| - | <li>pTet driving the expression of different genes</li> | + | <li>p<sub>Tet</sub> driving the expression of different genes</li> |
| | <ul><li>mRFP</li> | | <ul><li>mRFP</li> |
| | <ul class="disc"> | | <ul class="disc"> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516230 "> BBa_K516230 </a> pTet-RBS30-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516230 "> BBa_K516230 </a> p<sub>Tet</sub>-RBS30-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516231 "> BBa_K516231 </a> pTet-RBS31-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516231 "> BBa_K516231 </a> p<sub>Tet</sub>-RBS31-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516232 "> BBa_K516232 </a> pTet-RBS32-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516232 "> BBa_K516232 </a> p<sub>Tet</sub>-RBS32-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_I13521 "> BBa_I13521 </a> pTet-RBS34-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_I13521 "> BBa_I13521 </a> p<sub>Tet</sub>-RBS34-mRFP-TT </li> |
| | </ul> | | </ul> |
| | <li>AiiA</li> | | <li>AiiA</li> |
| | <ul class="disc"> | | <ul class="disc"> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516220 "> BBa_K516220 </a> pTet-RBS30-AiiA-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516220 "> BBa_K516220 </a> p<sub>Tet</sub>-RBS30-AiiA-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516221 "> BBa_K516221 </a> pTet-RBS31-AiiA-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516221 "> BBa_K516221 </a> p<sub>Tet</sub>-RBS31-AiiA-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516222 "> BBa_K516222 </a> pTet-RBS32-AiiA-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516222 "> BBa_K516222 </a> p<sub>Tet</sub>-RBS32-AiiA-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516224 "> BBa_K516224 </a> pTet-RBS34-AiiA-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516224 "> BBa_K516224 </a> p<sub>Tet</sub>-RBS34-AiiA-TT </li> |
| | </ul> | | </ul> |
| | <li>LuxI</li> | | <li>LuxI</li> |
| | <ul class="disc"> | | <ul class="disc"> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516210 "> BBa_K516210 </a> pTet-RBS30-LuxI </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516210 "> BBa_K516210 </a> p<sub>Tet</sub>-RBS30-LuxI </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516211 "> BBa_K516211 </a> pTet-RBS31-LuxI </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516211 "> BBa_K516211 </a> p<sub>Tet</sub>-RBS31-LuxI </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516212 "> BBa_K516212 </a> pTet-RBS32-LuxI </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516212 "> BBa_K516212 </a> p<sub>Tet</sub>-RBS32-LuxI </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516214 "> BBa_K516214 </a> pTet-RBS34-LuxI </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516214 "> BBa_K516214 </a> p<sub>Tet</sub>-RBS34-LuxI </li> |
| | </ul></ul> | | </ul></ul> |
| | </ul></ol> | | </ul></ol> |
| Line 380: |
Line 387: |
| | | | |
| | </p> | | </p> |
| | + | <p align='justify'> |
| | + | The results reported in the table suggest that the RBS efficiency ranking is not always maintained. In particular, for the different promoters driving the expression of mRFP, the ranking of the declared efficiencies is maintained for p<sub>Lux</sub>, but not for p<sub>Tet</sub> and J23101. The RBS B0030 results to be the most efficient for both J23101 and p<sub>Tet</sub>, but not for p<sub>Lux</sub> (NB: this effect might be due to an effective non-modularity of RBS, but also to saturating phenomena occurring for this very strong promoter at full induction). RBS B0031 always shows a very low efficiency, while B0032 an intermediate efficiency between B0031 and the stronger RBSs B0030 and B0034. <br> |
| | + | For what concerns the encoded gene variation, more significant differences can be observed. RBS B0030 has the higher efficiency only for mRFP, while the values for AiiA and LuxI are similar (~0.5). Unexpectedly, the weak RBS B0031 has a higher efficiency with AiiA gene (0.83), while with mRFP and LuxI. With B0032 no activity was observed for LuxI, while for AiiA and mRFP the results are quite consistent with the one reported above. |
| | + | </p> |
| | + | <p align='justify'> |
| | + | These results are encouraging: though the partial non-modularity of RBS with the encoded gene is confirmed, the hypothesis of modularity with the promoter is to some extent confirmed. Three classes of efficiencies were identified: |
| | + | <ul> |
| | + | <li>low efficiency RBS (B0031)</li> |
| | + | <li>medium efficiency RBS (B0032)</li> |
| | + | <li>high efficiency RBSs (B0030, B0034)</li> |
| | + | </ul> |
| | + | </p> |
| | + | |
| | | | |
| | | | |
| Line 394: |
Line 414: |
| | | | |
| | | | |
| - | <a name='pLux'></a><h2>pLux promoter</h2> | + | <a name='pLux'></a><h2>p<sub>Lux</sub> promoter</h2> |
| | <p align='justify'> | | <p align='justify'> |
| | | | |
| Line 402: |
Line 422: |
| | | | |
| | | | |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516330 "> BBa_K516330 </a> (wiki name: E17 ) pLambda-RBS30-LuxR-T-pLux-RBS30-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516330 "> BBa_K516330 </a> (wiki name: E17 ) pLambda-RBS30-LuxR-T-p<sub>Lux</sub>-RBS30-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516331 "> BBa_K516331 </a> (wiki name: E18 ) pLambda-RBS30-LuxR-T-pLux-RBS31-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516331 "> BBa_K516331 </a> (wiki name: E18 ) pLambda-RBS30-LuxR-T-p<sub>Lux</sub>-RBS31-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516332 "> BBa_K516332 </a> (wiki name: E19 ) pLambda-RBS30-LuxR-T-pLux-RBS32-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516332 "> BBa_K516332 </a> (wiki name: E19 ) pLambda-RBS30-LuxR-T-p<sub>Lux</sub>-RBS32-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516334 "> BBa_K516334 </a> (wiki name: E20 ) pLambda-RBS30-LuxR-T-pLux-RBS34-mRFP-TT </li></ul> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516334 "> BBa_K516334 </a> (wiki name: E20 ) pLambda-RBS30-LuxR-T-p<sub>Lux</sub>-RBS34-mRFP-TT </li></ul> |
| | </ol> | | </ol> |
| | | | |
| Line 449: |
Line 469: |
| | | | |
| | <div align="justify"> | | <div align="justify"> |
| - | <p>From this table, it is evident that, whilst α<sub>pLux</sub> assumes significantly different values for different RBSs, η<sub>pLux</sub> and k<sub>pLux</sub> assume very similar values. This result shows that RBS variation modulates the amplitude of Hill function, not affecting the shape of the curve. The four induction curves result to be the same Hill function modulated in amplitude by a parameter, that is the estimated RBS efficiency for this measurement system.</p> | + | <p>From this table, it is evident that, whilst α<sub>p<sub>Lux</sub></sub> assumes significantly different values for different RBSs, η<sub>pLux</sub> and k<sub>pLux</sub> assume very similar values. This result shows that RBS variation modulates the amplitude of Hill function, not affecting the shape of the curve. The four induction curves result to be the same Hill function modulated in amplitude by a parameter, that is the estimated RBS efficiency for this measurement system.</p> |
| | <p>These results are quite encouraging, because suggest that, given the non-modular behavior of RBS dpending on the encoded gene, the RBS has a modular behaviour respect to the promoter.</p> | | <p>These results are quite encouraging, because suggest that, given the non-modular behavior of RBS dpending on the encoded gene, the RBS has a modular behaviour respect to the promoter.</p> |
| | <p>The operative parameters are summarized in the table below:</p></div> | | <p>The operative parameters are summarized in the table below:</p></div> |
| Line 475: |
Line 495: |
| | </table> | | </table> |
| | <p align='justify'>These operative parameters provide useful information on the behavior of this 3OC6-HSL inducible device. RPU<sub>max</sub> assumes very different values in terms of RPUs. | | <p align='justify'>These operative parameters provide useful information on the behavior of this 3OC6-HSL inducible device. RPU<sub>max</sub> assumes very different values in terms of RPUs. |
| - | This can't be explained by RBS modulation, since RPUs have been evaluated by normalizing S<sub>cell</sub> of pLux-RBSx for the one of J23101-RBSx. It is evident that some nonlinear effect on maximum strength, maybe due to non-modularity of RBS when the promoter changes or maybe due to saturation effects on protein expression, occur. | + | This can't be explained by RBS modulation, since RPUs have been evaluated by normalizing S<sub>cell</sub> of p<sub>Lux</sub>-RBSx for the one of J23101-RBSx. It is evident that some nonlinear effect on maximum strength, maybe due to saturation phenomena on protein expression, occur. |
| - | The modulation in amplitude of the Hill can't be explained by a linear dependance on the RBS efficiency (in this case, in fact, the same RPUs should be observed for every RBS, since the standard reference used for RPUs computation). The switch point and linear boundaries are quite constant in all the cases, showing that the linear region of this system is not affected by RBS changes.</p> | + | The same RPUs should be observed for every RBS, since the normalization by the standard reference used for RPUs computation should eliminate the RBS contribute. Here different RPUs are observed, maybe due to nonlinear RBS behavior or to saturation phenomena occurring with this very strong promoter. The switch point and linear boundaries are quite constant in all the cases, showing that the linear region of this system is not affected by RBS changes.</p> |
| | | | |
| | <table width='100%'><tr><td width='50%'> | | <table width='100%'><tr><td width='50%'> |
| Line 501: |
Line 521: |
| | <!-- ----------------- --> | | <!-- ----------------- --> |
| | | | |
| - | <a name='pTet'></a><h2>pTet promoter</h2> | + | <a name='pTet'></a><h2>p<sub>Tet</sub> promoter</h2> |
| | | | |
| | <ol> | | <ol> |
| | <ul> | | <ul> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516230 "> BBa_K516230 </a> (wiki name: E21 ) pTet-RBS30-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516230 "> BBa_K516230 </a> (wiki name: E21 ) p<sub>Tet</sub>-RBS30-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516231 "> BBa_K516231 </a> (wiki name: E22 ) pTet-RBS31-mRFP-TT </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516231 "> BBa_K516231 </a> (wiki name: E22 ) p<sub>Tet</sub>-RBS31-mRFP-TT </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516232 "> BBa_K516232 </a> (wiki name: E23 ) pTet-RBS32-mRFP-TT | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516232 "> BBa_K516232 </a> (wiki name: E23 ) p<sub>Tet</sub>-RBS32-mRFP-TT |
| | </li> | | </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_I13521 "> BBa_I13521 </a> pTet-RBS34-mRFP-TT | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_I13521 "> BBa_I13521 </a> p<sub>Tet</sub>-RBS34-mRFP-TT |
| | </li></ul> | | </li></ul> |
| | </ul> | | </ul> |
| Line 515: |
Line 535: |
| | | | |
| | <div align="justify"> | | <div align="justify"> |
| - | <p>The protocols for the characterization of pTet promoter are reported in the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#pTet_protocol'>pTet measurement section</a>.</p> | + | <p>The protocols for the characterization of p<sub>Tet<sub> promoter are reported in the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#pTet_protocol'>p<sub>Tet</sub> measurement section</a>.</p> |
| | + | <p>The data collected from the mRFP measurement systems were processed as described in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements'> data analysis section</a>. The induction curves were obtained by fitting a Hill function as described in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Ptet_&_Plux'>modelling section</a> and the estimated <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Table_of_parameters'>parameters</a> for p<sub>Tet</sub> are reported in the pictures and in table below. </p> |
| | <p>This promoter is widely studied and characterized usually using the strong RBS BBa_B0034. Here we have characterized its transcriptional strength as a function of aTc induction (ng/ul) for different RBSs. Three different induction curves were obtained and are reported in figure:</p></div> | | <p>This promoter is widely studied and characterized usually using the strong RBS BBa_B0034. Here we have characterized its transcriptional strength as a function of aTc induction (ng/ul) for different RBSs. Three different induction curves were obtained and are reported in figure:</p></div> |
| | | | |
| Line 526: |
Line 547: |
| | <div style="text-align:justify"><div class="thumbinner" width='100%'><a href="https://static.igem.org/mediawiki/2011/9/93/E33_RPU_80.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/9/93/E33_RPU_80.jpg" class="thumbimage" width="100%"></a></div></div> | | <div style="text-align:justify"><div class="thumbinner" width='100%'><a href="https://static.igem.org/mediawiki/2011/9/93/E33_RPU_80.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/9/93/E33_RPU_80.jpg" class="thumbimage" width="100%"></a></div></div> |
| | </td> --> | | </td> --> |
| - |
| |
| | | | |
| | <table width='100%'> | | <table width='100%'> |
| Line 537: |
Line 557: |
| | </table> | | </table> |
| | | | |
| - | <p>The data collected from the mRFP measurement systems were processed as described in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements'> data analysis section</a>. The induction curves were obtained by fitting a Hill function as described in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Ptet_&_Plux'>modelling section</a> and the estimated <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Table_of_parameters'>parameters</a> for pTet are reported in the pictures and in table below. </p>
| + | |
| | <p align='justify'> | | <p align='justify'> |
| | The estimated parameters of the Hill curves described in the figures are summarized in the table below: | | The estimated parameters of the Hill curves described in the figures are summarized in the table below: |
| Line 579: |
Line 599: |
| | | | |
| | <p align='justify'> | | <p align='justify'> |
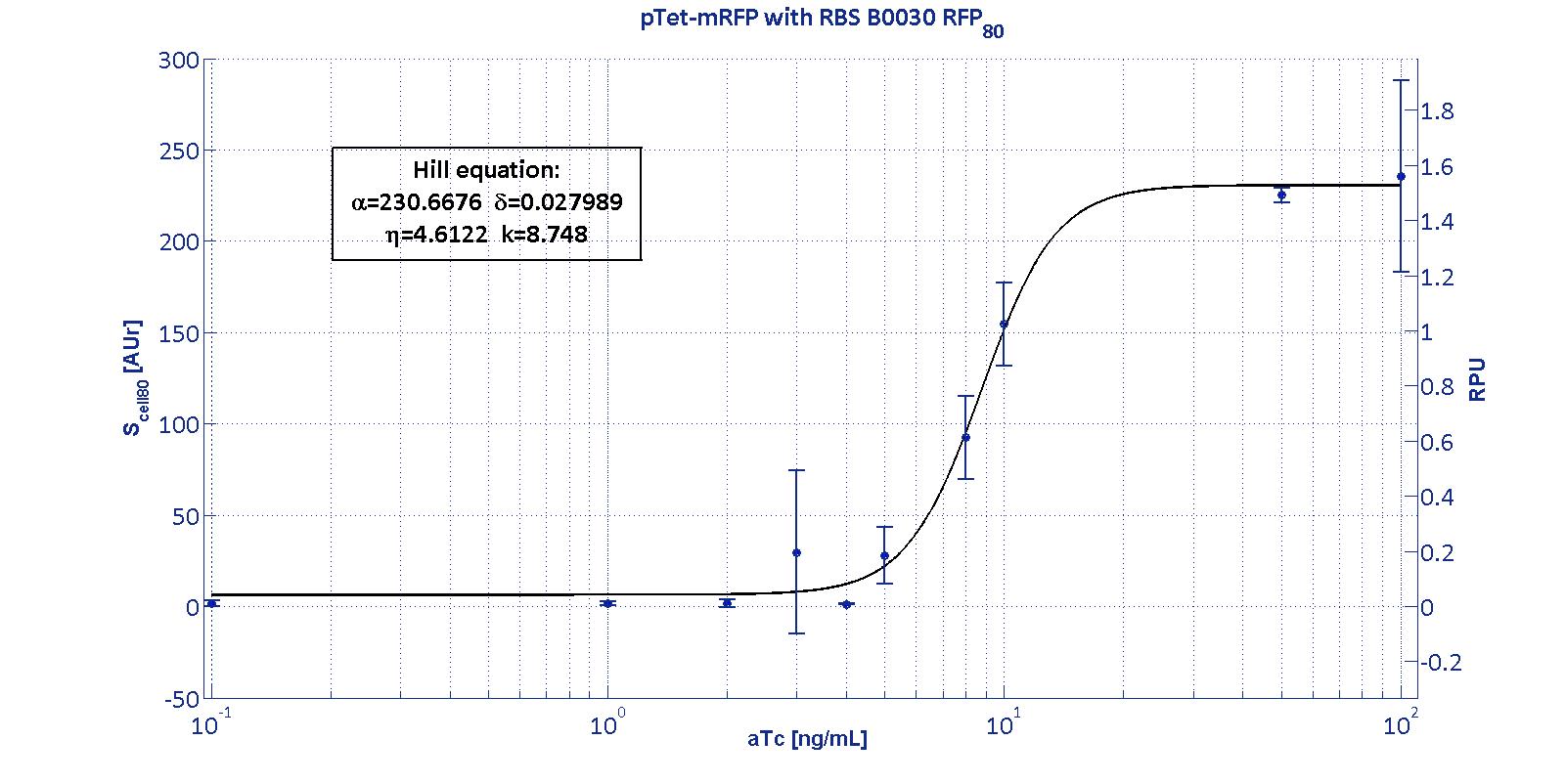
| - | The measurement system pTet-B0031-mRFP-TT couldn't be assayed because its fluorescence output is under the detectability threshold of our measurement instrument. For this reason, the parameters of the corresponding Hill curve couldn't be estimated and are reported as 'Not Determined' ND. | + | The measurement system p<sub>Tet</sub>-B0031-mRFP-TT couldn't be assayed because its fluorescence output is under the detectability threshold of our measurement instrument. For this reason, the parameters of the corresponding Hill curve couldn't be estimated and are reported as 'Not Determined' ND. |
| | </p> | | </p> |
| | | | |
| | <div align='justify'> | | <div align='justify'> |
| | <p>α parameter (representing the maximum trascriptional rate in the studied range of induction) varies as expected with the RBS variation and also the δ and η parameters are quite different among the RBS variations.</p> | | <p>α parameter (representing the maximum trascriptional rate in the studied range of induction) varies as expected with the RBS variation and also the δ and η parameters are quite different among the RBS variations.</p> |
| - | <p>The k<sub>pTet</sub> parameter is quite constant among the RBS variations, thus suggesting that in this case the RBS variation doesn't substantially affect the switch point of the Hill curve, even if the amplitude and the maximum slope are not quite maintained (for the η parameter, maybe fitting problems). | + | <p>The k<sub>p<sub>Tet</sub></sub> parameter is quite constant among the RBS variations, thus suggesting that in this case the RBS variation doesn't substantially affect the switch point of the Hill curve, even if the amplitude and the maximum slope are not quite maintained (for the η parameter, maybe fitting problems). |
| | | | |
| | </p> | | </p> |
| Line 635: |
Line 655: |
| | <p align='justify'> | | <p align='justify'> |
| | | | |
| - | LuxI has been characterized through the Biosensor BBa_T9002 (see <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#introduction_to_T9002'>modelling section</a>) using the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#pTetLuxI'>pTet-RBSx-LuxI-TT</a> measurement systems. | + | LuxI has been characterized through the Biosensor BBa_T9002 (see <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#introduction_to_T9002'>modelling section</a>) using the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#pTetLuxI'>p<sub>Tet</sub>-RBSx-LuxI-TT</a> measurement systems. |
| | <br>The HSL synthesis rate has been evaluated according to the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Equations_for_gene_networks'>model equations</a>, properly adjusted. In particular, the ODE system is reported here: </p> | | <br>The HSL synthesis rate has been evaluated according to the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Equations_for_gene_networks'>model equations</a>, properly adjusted. In particular, the ODE system is reported here: </p> |
| | | | |
| Line 645: |
Line 665: |
| | | | |
| | <p align='justify'> | | <p align='justify'> |
| - | Since the measurement systems are only assayed in the exponential growth phase, in the equation (3) N<<N<sub>max</sub> is assumed. | + | Since the measurement systems are only assayed in the exponential growth phase, in the equation (3) N<<N<sub>max</sub> is assumed. |
| | The parameters N<sub>max</sub>, μ and γ<sub>HSL</sub> (see the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Results'>Results section</a> for more details) are known and summarized in the table below:</p> | | The parameters N<sub>max</sub>, μ and γ<sub>HSL</sub> (see the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Results'>Results section</a> for more details) are known and summarized in the table below:</p> |
| | | | |
| Line 664: |
Line 684: |
| | | | |
| | <p align='justify'> | | <p align='justify'> |
| - | The parameters V<sub>max</sub>, k<sub>M,LuxI</sub> and α<sub>RBSx</sub> were estimated with a simultaneous fitting of the data collected as described in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#LuxI'>measurement section</a> for the four measurement parts <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#pTetLuxI'>pTet-RBSx-LuxI-TT</a> assayed by <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#T9002'>BBa_T9002 biosensor</a> section. | + | The parameters V<sub>max</sub>, k<sub>M,LuxI</sub> and α<sub>RBSx</sub> were estimated with a simultaneous fitting of the data collected as described in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#LuxI'>measurement section</a> for the four measurement parts <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#pTetLuxI'>p<sub>Tet</sub>-RBSx-LuxI-TT</a> assayed by <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#T9002'>BBa_T9002 biosensor</a> section. |
| | | | |
| | | | |
| Line 687: |
Line 707: |
| | <tr><td class='row'>3.56*10<sup>-9</sup></td> | | <tr><td class='row'>3.56*10<sup>-9</sup></td> |
| | <td class='row'>6.87*10<sup>3</sup></td> | | <td class='row'>6.87*10<sup>3</sup></td> |
| - | <td class='row'>252</td> | + | <td class='row'> 87 </td> |
| | <td class='row'>8.5</td> | | <td class='row'>8.5</td> |
| | <td class='row'>ND</td> | | <td class='row'>ND</td> |
| - | <td class='row'>87</td> | + | <td class='row'> 252 </td> |
| | </tr></table> | | </tr></table> |
| | </div> | | </div> |
| Line 718: |
Line 738: |
| | | | |
| | <p align='justify'> | | <p align='justify'> |
| - | The activity of AiiA enzyme has been evaluated by testing the measurement systems <a href="https://static.igem.org/mediawiki/2011/1/11/Pc_aiia.jpg">pTet-RBSx-AiiA-TT</a>. | + | The activity of AiiA enzyme has been evaluated by testing the measurement systems <a href="https://static.igem.org/mediawiki/2011/1/11/Pc_aiia.jpg">p<sub>Tet</sub>-RBSx-AiiA-TT</a>. |
| | Similar to LuxI, a system of differential equations (referring to <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Equations_for_gene_networks'>model equations</a>) has been derived. | | Similar to LuxI, a system of differential equations (referring to <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Equations_for_gene_networks'>model equations</a>) has been derived. |
| | </p> | | </p> |
| Line 729: |
Line 749: |
| | | | |
| | <p align='justify'> | | <p align='justify'> |
| - | The parameters k<sub>cat</sub>, k<sub>M,AiiA</sub> and α<sub>RBSx</sub> would have been estimated with a simultaneous fitting of the data collected as described in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#AiiA'>measurement section</a> for the four measurement parts <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#pTetAiiA'>pTet-RBSx-AiiA-TT</a> assayed by <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#T9002'>BBa_T9002 biosensor</a> section. | + | The parameters k<sub>cat</sub>, k<sub>M,AiiA</sub> and α<sub>RBSx</sub> would have been estimated with a simultaneous fitting of the data collected as described in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#AiiA'>measurement section</a> for the four measurement parts <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#pTetAiiA'>p<sub>Tet</sub>-RBSx-AiiA-TT</a> assayed by <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#T9002'>BBa_T9002 biosensor</a> section. |
| | Unfortunately, their estimation revealed impossible.<br> | | Unfortunately, their estimation revealed impossible.<br> |
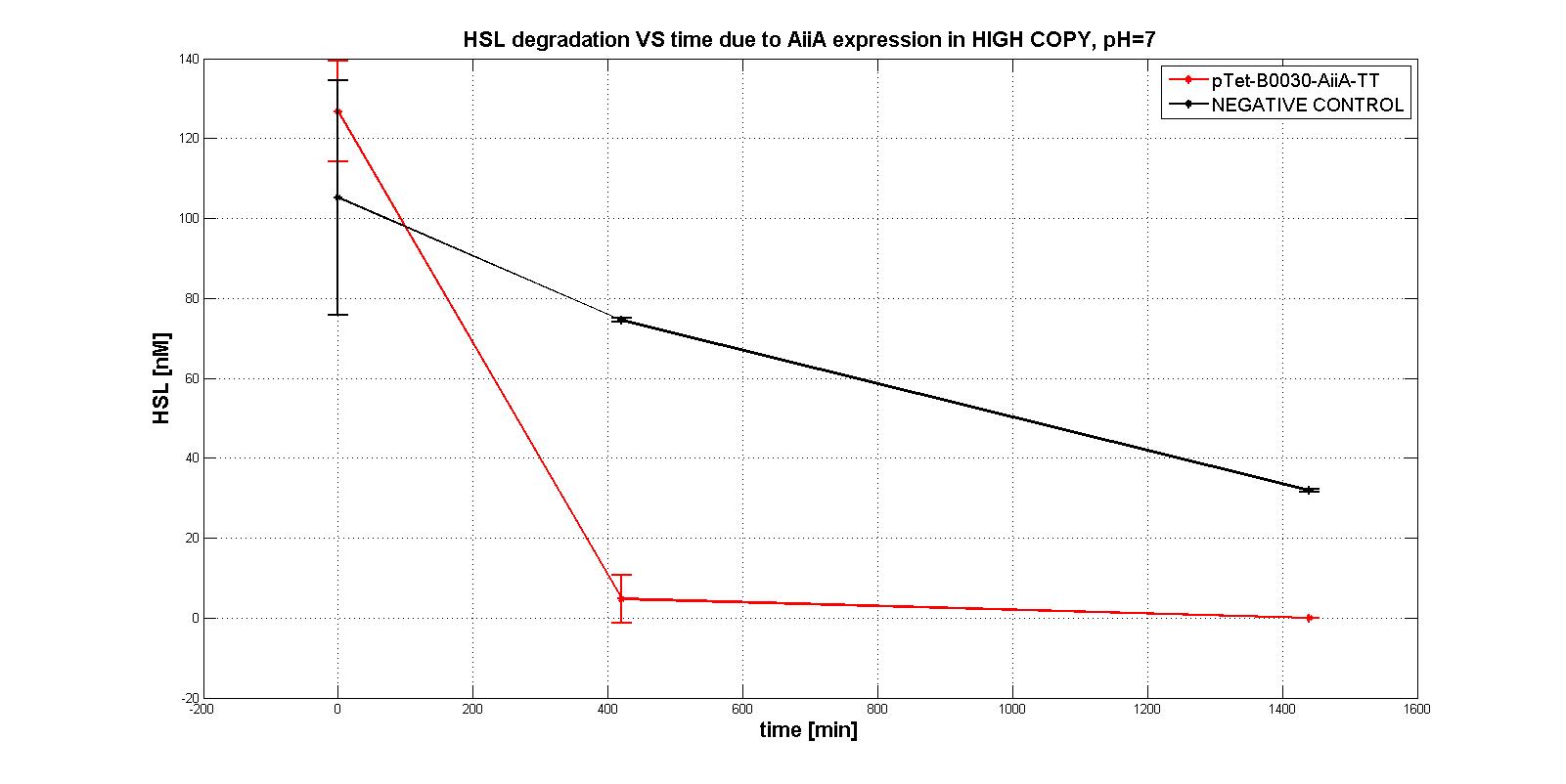
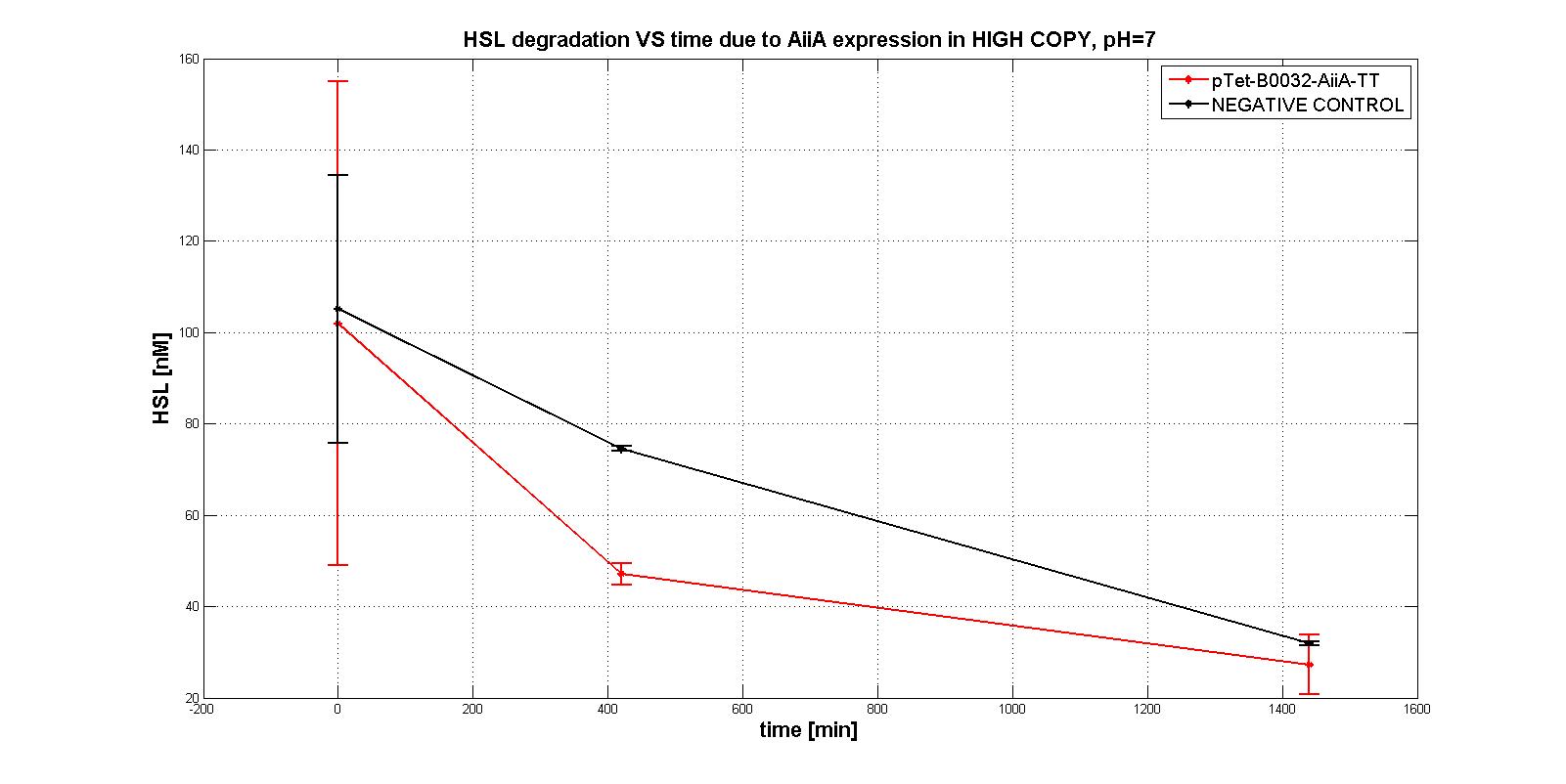
| - | Firstly several tests were performed, considering the subpart <a href="https://static.igem.org/mediawiki/2011/1/11/Pc_aiia.jpg">pTet-RBSx-AiiA-TT</a> in LOW-COPY at pH=7. An example of them is shown in the figure below: HSL is degraded in time as well as the negative control, which shouldn't degrade it even after 21 hours. | + | Firstly several tests were performed, considering the subpart <a href="https://static.igem.org/mediawiki/2011/1/11/Pc_aiia.jpg">p<sub>Tet</sub>-RBSx-AiiA-TT</a> in LOW-COPY at pH=7. An example of them is shown in the figure below: HSL is degraded in time as well as the negative control, which shouldn't degrade it even after 21 hours. |
| | | | |
| | | | |
| Line 761: |
Line 781: |
| | | | |
| | | | |
| - | <div align="justify">So, we decided to investigate the behaviour in HIGH COPY in another <em>E.COLI</em> strain, TOP10, in order to understand if the enzyme AiiA would have been worked well. In this case, a significant difference in degradation between pTet-RBSx-AiiA-TT and the negative control was observed, also just after 7 hours. That was the proof of the good functioning (in HIGH COPY) of the enzyme.</div> | + | <div align="justify">So, we decided to investigate the behaviour in HIGH COPY in another <em>E.COLI</em> strain, TOP10, in order to understand if the enzyme AiiA would have been worked well. In this case, a significant difference in degradation between p<sub>Tet</sub>-RBSx-AiiA-TT and the negative control was observed, also just after 7 hours. That was the proof of the good functioning (in HIGH COPY) of the enzyme.</div> |
| | | | |
| | | | |
| Line 830: |
Line 850: |
| | <!-- ----------------- --> | | <!-- ----------------- --> |
| | | | |
| - | <a name='K999'></a><h2>pSB1C3 plasmid with mRFP between S and P bearing pTet (easy-to-clone)</h2> | + | <a name='K999'></a><h2>pSB1C3 plasmid with mRFP between S and P bearing p<sub>Tet</sub> (easy-to-clone)</h2> |
| | <table align='center' width='100%'> | | <table align='center' width='100%'> |
| | <tr> | | <tr> |
| Line 839: |
Line 859: |
| | </tr> | | </tr> |
| | </table> | | </table> |
| - | <div align="justify">This vector was designed and realized in order to facilitate the cloning of coding sequences downstream of the strong promoter pTet. This vector was assembled by ligating S-P excided mRFP coding sequence from BBa_J61002 and ligating it in BBa_R0040 cut with S and P. Thus, the resulting vector contains mRFP between S and P. pTet cn be easily excided (E-P) and moved in the desired vector (E-P) and then the desired coding sequence can be easily assembled by digesting S-P the vector and X-P the coding sequence, thus obtaining a final part that is standard10-compatible.</div><br> | + | <div align="justify">This vector was designed and realized in order to facilitate the cloning of coding sequences downstream of the strong promoter p<sub>Tet</sub>. This vector was assembled by ligating S-P excided mRFP coding sequence from BBa_J61002 and ligating it in BBa_R0040 cut with S and P. Thus, the resulting vector contains mRFP between S and P. p<sub>Tet</sub> can be easily excided (E-P) and moved in the desired vector (E-P) and then the desired coding sequence can be easily assembled by digesting S-P the vector and X-P the coding sequence, thus obtaining a final part that is standard10-compatible.</div><br> |
| | | | |
| | <div align="right"><small><a href="#indice" title="">^top</a></small></div> | | <div align="right"><small><a href="#indice" title="">^top</a></small></div> |
| Line 853: |
Line 873: |
| | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516030 "> BBa_K516030 </a> (wiki name: E5 ) RBS30-mRFP-TT Rebuilt existing part from BBa_S04180 (DNA planning) </li> | | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516030 "> BBa_K516030 </a> (wiki name: E5 ) RBS30-mRFP-TT Rebuilt existing part from BBa_S04180 (DNA planning) </li> |
| | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516032 "> BBa_K516032 </a> (wiki name: E7 ) RBS32-mRFP-TT Rebuilt existing part from BBa_ J133000 (DNA planning) </li> | | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516032 "> BBa_K516032 </a> (wiki name: E7 ) RBS32-mRFP-TT Rebuilt existing part from BBa_ J133000 (DNA planning) </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516214 "> BBa_K516214 </a> (wiki name: E16 ) pTet-RBS34-LuxI Rebuilt existing part from BBa_S03623 (DNA available, only 2008 kit, inconsistent) </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516214 "> BBa_K516214 </a> (wiki name: E16 ) p<sub>Tet</sub>-RBS34-LuxI Rebuilt existing part from BBa_S03623 (DNA available, only 2008 kit, inconsistent) </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516222 "> BBa_K516222 </a> (wiki name: E26 ) pTet-RBS32-AiiA-TT Rebuilt existing part from BBa_ J22071 (2008 only, Bad sequencing) </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516222 "> BBa_K516222 </a> (wiki name: E26 ) p<sub>Tet</sub>-RBS32-AiiA-TT Rebuilt existing part from BBa_ J22071 (2008 only, Bad sequencing) </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516224 "> BBa_K516224 </a> (wiki name: E27 ) pTet-RBS34-AiiA-TT Rebuilt existing part from BBa_K077047 (Part deleted) </li> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516224 "> BBa_K516224 </a> (wiki name: E27 ) p<sub>Tet</sub>-RBS34-AiiA-TT Rebuilt existing part from BBa_K077047 (Part deleted) </li> |
| - | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516232 "> BBa_K516232 </a> (wiki name: E23 ) pTet-RBS32-mRFP-TT Rebuilt existing part from BBa_I20252 (DNA planning) </li></ul></ol> | + | <li> <A HREF="http://partsregistry.org/wiki/index.php/Part: BBa_K516232 "> BBa_K516232 </a> (wiki name: E23 ) p<sub>Tet</sub>-RBS32-mRFP-TT Rebuilt existing part from BBa_I20252 (DNA planning) </li></ul></ol> |
| | </div> | | </div> |
| | | | |


 "
"