Team:ETH Zurich
From 2011.igem.org
| (21 intermediate revisions not shown) | |||
| Line 11: | Line 11: | ||
<div><img src="/wiki/images/d/d6/ETHZ_FRONT_Slide_2.png" usemap="#Slide2"/> | <div><img src="/wiki/images/d/d6/ETHZ_FRONT_Slide_2.png" usemap="#Slide2"/> | ||
<map name="Slide2"> | <map name="Slide2"> | ||
| - | <area shape="circle" alt="" title="" coords=" | + | <area shape="circle" alt="" title="" coords="320,150,88" href="/Team:ETH_Zurich/Overview/Informationprocessing#Signal_Pre-Processing" /> |
| - | <area shape="circle" alt="" title="" coords="256,272,68" href="/Team:ETH_Zurich/Overview/ | + | <area shape="circle" alt="" title="" coords="256,272,68" href="/Team:ETH_Zurich/Overview/Informationprocessing#Signal_Detection" /> |
| - | <area shape="circle" alt="" title="" coords=" | + | <area shape="circle" alt="" title="" coords="330,372,88" href="/Team:ETH_Zurich/Overview/Informationprocessing#I:_The_Band-Pass_Filter" /> |
| - | <area shape="circle" alt="" title="" coords="197,421,65" href="/Team:ETH_Zurich/Overview/ | + | <area shape="circle" alt="" title="" coords="197,421,65" href="/Team:ETH_Zurich/Overview/Informationprocessing#II:_The_Alarm_Signal" /> |
</map> | </map> | ||
</div> | </div> | ||
| Line 20: | Line 20: | ||
<div><img src="/wiki/images/4/46/ETHZ_FRONT_Slide_3.png" usemap="#Slide3" /> | <div><img src="/wiki/images/4/46/ETHZ_FRONT_Slide_3.png" usemap="#Slide3" /> | ||
<map name="Slide3"> | <map name="Slide3"> | ||
| - | + | <area shape="circle" alt="" title="" coords="181,181,103" href="/Team:ETH_Zurich/Process/Validation" /> | |
| + | <area shape="circle" alt="" title="" coords="346,276,90" href="/Team:ETH_Zurich/Process/Microfluidics/Proof#Experimental_setup" /> | ||
| + | <area shape="circle" alt="" title="" coords="493,177,97" href="/Team:ETH_Zurich/Process/Validation#Setup_Validation" /> | ||
| + | <area shape="circle" alt="" title="" coords="635,259,93" href="/Team:ETH_Zurich/Process/Microfluidics/Proof#Results"/> | ||
</map> | </map> | ||
</div> | </div> | ||
| Line 26: | Line 29: | ||
<div><img src="/wiki/images/d/d9/ETHZ_FRONT_Slide_4.png" usemap="#Slide4"/> | <div><img src="/wiki/images/d/d9/ETHZ_FRONT_Slide_4.png" usemap="#Slide4"/> | ||
<map name="Slide4"> | <map name="Slide4"> | ||
| - | <area shape="circle" title="" coords="111,157,83" href="" /> | + | <area shape="circle" title="" coords="111,157,83" href="/Team:ETH_Zurich/Process/Validation#Setup_Validation" /> |
| - | <area shape="circle" title="" coords="251,233,81" href="/Team:ETH_Zurich/Biology/Validation# | + | <area shape="circle" title="" coords="251,233,81" href="/Team:ETH_Zurich/Biology/Validation##XylR_-_the_xylene_sensor" /> |
<area shape="circle" title="" coords="387,147,79" href="/Team:ETH_Zurich/Achievements/Data_page" /> | <area shape="circle" title="" coords="387,147,79" href="/Team:ETH_Zurich/Achievements/Data_page" /> | ||
<area shape="circle" title="" coords="524,230,80" href="/Team:ETH_Zurich/Biology/Validation#Alarm" /> | <area shape="circle" title="" coords="524,230,80" href="/Team:ETH_Zurich/Biology/Validation#Alarm" /> | ||
| - | <area shape="circle" title="" coords="663,149,85" href="" /> | + | <area shape="circle" title="" coords="663,149,85" href="/Team:ETH_Zurich/Biology/Validation" /> |
</map> | </map> | ||
</div> | </div> | ||
| Line 39: | Line 42: | ||
<area shape="circle" alt="" title="" coords="308,252,64" href="/Team:ETH_Zurich/Modeling/SingleCell" target="" /> | <area shape="circle" alt="" title="" coords="308,252,64" href="/Team:ETH_Zurich/Modeling/SingleCell" target="" /> | ||
<area shape="circle" alt="" title="" coords="366,370,70" href="/Team:ETH_Zurich/Modeling/Combined" target="" /> | <area shape="circle" alt="" title="" coords="366,370,70" href="/Team:ETH_Zurich/Modeling/Combined" target="" /> | ||
| + | <area shape="circle" alt="" title="" coords="130,268,78" href="/Team:ETH_Zurich/Modeling/Stochastic" target="" /> | ||
| + | <area shape="circle" alt="" title="" coords="542,110,78" href="/Team:ETH_Zurich/Modeling/Analytical_Approximation" /> | ||
</map> | </map> | ||
</div> | </div> | ||
| Line 47: | Line 52: | ||
<area shape="rect" alt="" title="" coords="37,110,317,187" href="/Team:ETH_Zurich/Team/Acknowledgements" /> | <area shape="rect" alt="" title="" coords="37,110,317,187" href="/Team:ETH_Zurich/Team/Acknowledgements" /> | ||
<area shape="rect" alt="" title="" coords="337,75,450,137" href="/Team:ETH_Zurich/Biology/Safety" /> | <area shape="rect" alt="" title="" coords="337,75,450,137" href="/Team:ETH_Zurich/Biology/Safety" /> | ||
| - | <area shape="rect" alt="" title="" coords="522,16,764,86" href="/Team:ETH_Zurich/Team/Human_practices#22.10. | + | <area shape="rect" alt="" title="" coords="522,16,764,86" href="/Team:ETH_Zurich/Team/Human_practices#22.10.11_D-BSSE_Open_house" /> |
| - | <area shape="rect" alt="" title="" coords="518,117,758,190" href="/Team:ETH_Zurich/Team/Human_practices#7.10. | + | <area shape="rect" alt="" title="" coords="518,117,758,190" href="/Team:ETH_Zurich/Team/Human_practices#7.10.11_CBB_Get_together_afternoon" /> |
</map> | </map> | ||
</div> | </div> | ||
| Line 55: | Line 60: | ||
{{:Team:ETH Zurich/Templates/SectionStart}} | {{:Team:ETH Zurich/Templates/SectionStart}} | ||
| - | = Abstract = | + | = SmoColi - Abstract = |
| - | '''We create a bacterio-quantifier of smoke. SmoColi cells are engineered to sense toxic substances found in cigarette smoke. They are immobilized in a microfluidic channel, in which a concentration gradient of the toxic substance is established. The sensor is linked to a band-pass filter that leads to input-concentration-dependent GFP expression. Continuous increase of the input concentration and its detection, therefore, establishes a moving fluorescent band in the channel. Finally, if the input concentration exceeds a certain threshold, cells produce RFP and the device turns red.''' | + | '''We create a bacterio-quantifier of smoke. SmoColi cells are engineered to sense toxic substances found in cigarette smoke (for example acetaldehyde or xylene). They are immobilized in a microfluidic channel, in which a concentration gradient of the toxic substance is established. The sensor is linked to a band-pass filter that leads to input-concentration-dependent GFP expression. Continuous increase of the input concentration and its detection, therefore, establishes a moving fluorescent band in the channel. Finally, if the input concentration exceeds a certain threshold, cells produce RFP and the device turns red. Due to its modularity, our SmoColi system can be used in fact as a quantifier for a range of substances, as long as the sensor is adapted. To show that our system can be activated by different signals, as a proof of principle, we modified the circuit so that it can be induced by arabinose.''' |
{{:Team:ETH Zurich/Templates/SectionEnd}} | {{:Team:ETH Zurich/Templates/SectionEnd}} | ||
{{:Team:ETH Zurich/Templates/HeaderNewEnd}} | {{:Team:ETH Zurich/Templates/HeaderNewEnd}} | ||
Latest revision as of 01:31, 29 October 2011




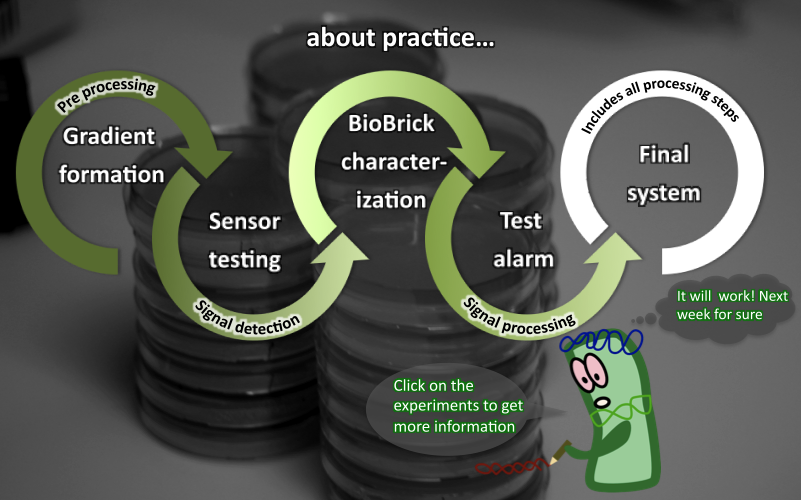
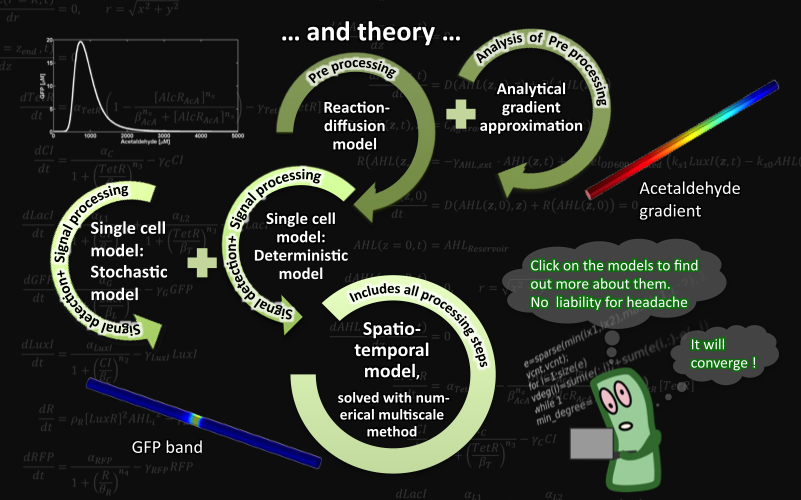

SmoColi - Abstract
We create a bacterio-quantifier of smoke. SmoColi cells are engineered to sense toxic substances found in cigarette smoke (for example acetaldehyde or xylene). They are immobilized in a microfluidic channel, in which a concentration gradient of the toxic substance is established. The sensor is linked to a band-pass filter that leads to input-concentration-dependent GFP expression. Continuous increase of the input concentration and its detection, therefore, establishes a moving fluorescent band in the channel. Finally, if the input concentration exceeds a certain threshold, cells produce RFP and the device turns red. Due to its modularity, our SmoColi system can be used in fact as a quantifier for a range of substances, as long as the sensor is adapted. To show that our system can be activated by different signals, as a proof of principle, we modified the circuit so that it can be induced by arabinose.
 "
"




