Team:DTU-Denmark/Project
From 2011.igem.org
(→Results) |
|||
| (57 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
<html> | <html> | ||
| - | <iframe style="float: right; margin: | + | <iframe style="float: right; margin: 15px 0 20px 10px; border: 4px solid white; outline: 1px solid #CCCCCC;" width="440" height="350" src="http://www.youtube.com/embed/dtleYPzOg-Y" frameborder="0" allowfullscreen></iframe> |
</html> | </html> | ||
| Line 14: | Line 14: | ||
== Abstract == | == Abstract == | ||
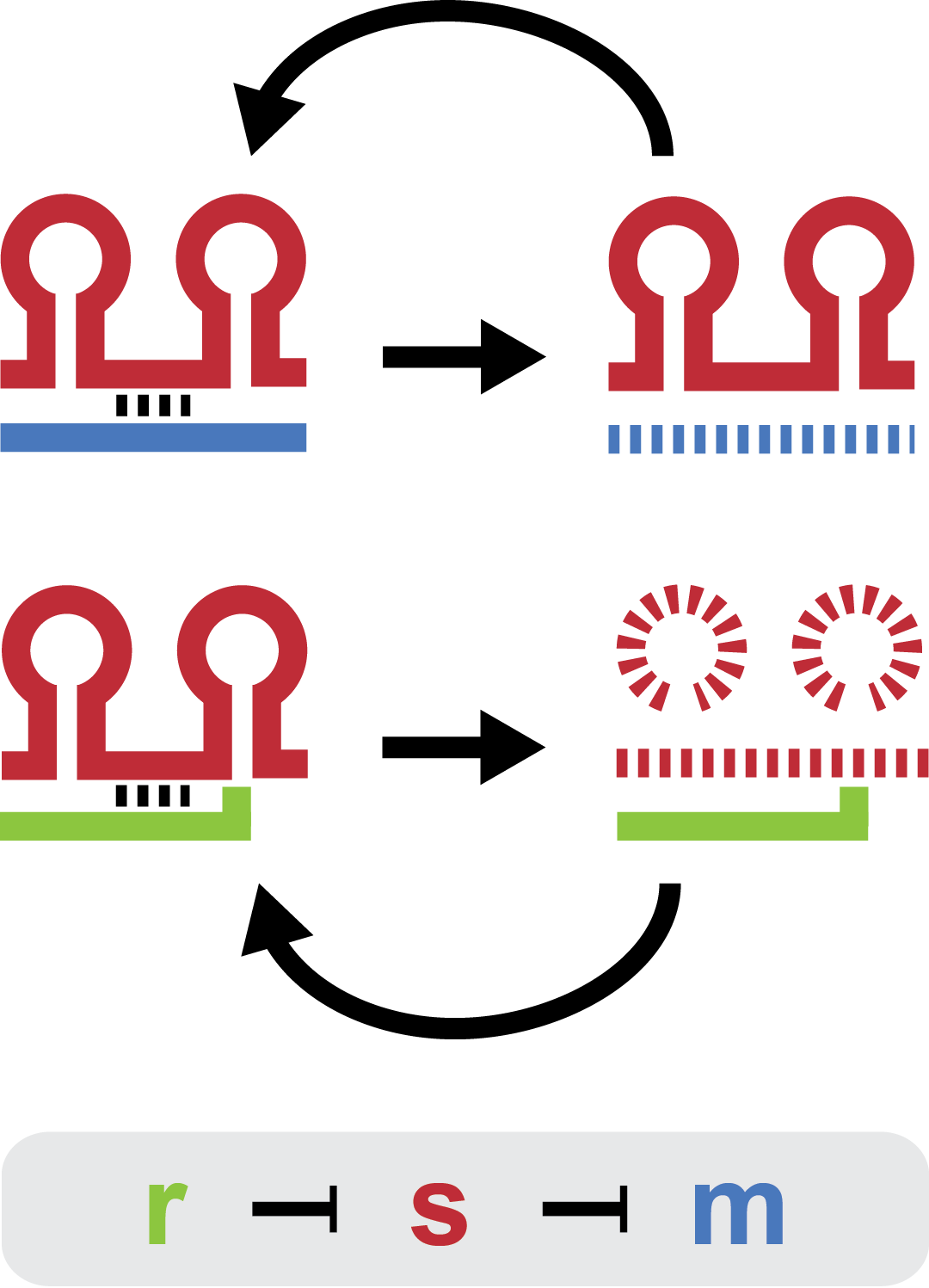
| - | Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. Such applications include convenient gene silencing and fine-tuning of gene expression, which are currently cumbersome processes restricted to well studied bacteria. We have investigated a novel type of RNA regulation based on the [[Team:DTU-Denmark/Background_the_natural_system|chitobiose system]], where the inhibition caused by a small RNA is relieved by another small RNA called trap-RNA.[[File:DTU2011_project_fig1.png|100px|frameless| | + | Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. Such applications include convenient gene silencing and fine-tuning of gene expression, which are currently cumbersome processes restricted to well studied bacteria. We have investigated a novel type of RNA regulation based on the [[Team:DTU-Denmark/Background_the_natural_system|chitobiose system]], where the inhibition caused by a small RNA is relieved by another small RNA called trap-RNA.[[File:DTU2011_project_fig1.png|100px|frameless|right|Two-level sRNA regulation. Blue is any target mRNA, red is sRNA and green is trap-RNA.]] We explore the possibility of using the system to uniquely target and repress any gene of interest, potentially providing unprecedented specificity and control of gene silencing. We furthermore constructed araBAD promoters with varying promoter activities using synthetic promoter libraries. |
</div> | </div> | ||
| Line 23: | Line 23: | ||
== Experiment: Testing sRNA == | == Experiment: Testing sRNA == | ||
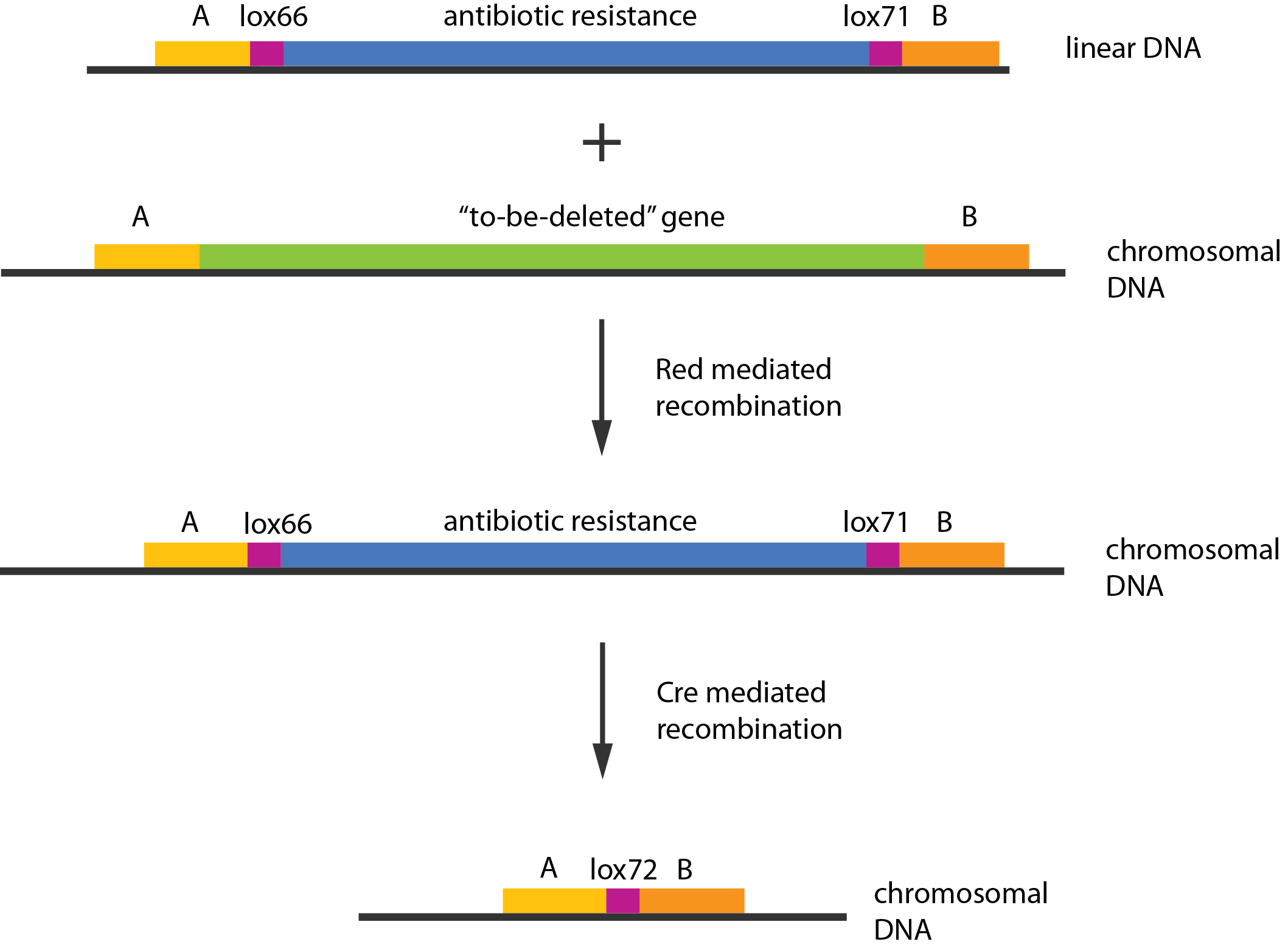
| - | [[File:DTU1_Redswap.png|200px|frameless| | + | [[File:DTU1_Redswap.png|200px|frameless|right|link=Team:DTU-Denmark/Project_experiment|Chromosomal knockout]] |
| - | + | ||
| - | + | ||
| + | Experiments were performed to verify that the envisioned small RNA based gene silencing is possible. Plasmids containing and strains deleted for the components were constructed providing a biological model. | ||
[[Team:DTU-Denmark/Project_experiment|Read more...]] | [[Team:DTU-Denmark/Project_experiment|Read more...]] | ||
| Line 34: | Line 33: | ||
== Bioinformatics == | == Bioinformatics == | ||
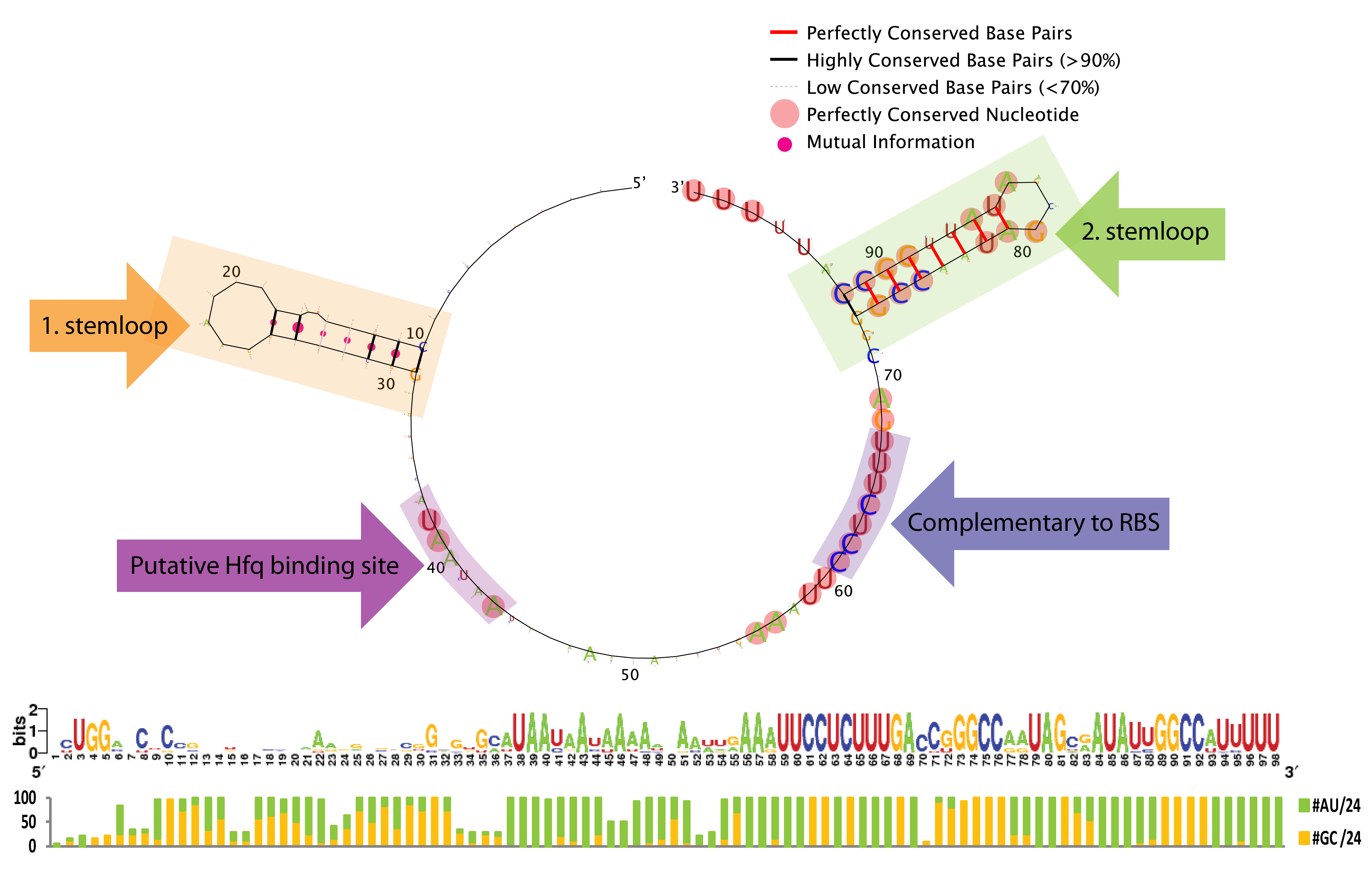
| - | [[File:DTU1 Sequence logo.png|200px|frameless| | + | [[File:DTU1 Sequence logo.png|200px|frameless|right|link=Team:DTU-Denmark/Bioinformatic]] |
A bioinformatics study was performed to investigate the possibilities of engineering the trap-RNA system to target any gene. The study elucidates interesting features of sequence and secondary structure conservation guiding future application. | A bioinformatics study was performed to investigate the possibilities of engineering the trap-RNA system to target any gene. The study elucidates interesting features of sequence and secondary structure conservation guiding future application. | ||
| - | |||
[[Team:DTU-Denmark/Bioinformatic|Read more...]] | [[Team:DTU-Denmark/Bioinformatic|Read more...]] | ||
| Line 47: | Line 45: | ||
== Modeling == | == Modeling == | ||
| - | [[File:DTU2011_modeling_fig.png|200px|frameless|right|'''Kinetic models''' of the system are the basis for modeling. Blue is target mRNA, red is small RNA and green is trap-RNA]] | + | [[File:DTU2011_modeling_fig.png|200px|frameless|right|link=Team:DTU-Denmark/Modeling|'''Kinetic models''' of the system are the basis for modeling. Blue is target mRNA, red is small RNA and green is trap-RNA]] |
| - | + | ||
| - | + | ||
| + | A framework for characterization of gene silencing was developed to guide rational design and test hypotheses. Steady state analysis revealed that each trap-RNA system has a characteristic fold repression. | ||
[[Team:DTU-Denmark/Modeling|Read more...]] | [[Team:DTU-Denmark/Modeling|Read more...]] | ||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
| Line 60: | Line 54: | ||
<div class="overviewBox left"> | <div class="overviewBox left"> | ||
| - | == Experiment: | + | == Experiment: Improving araBAD == |
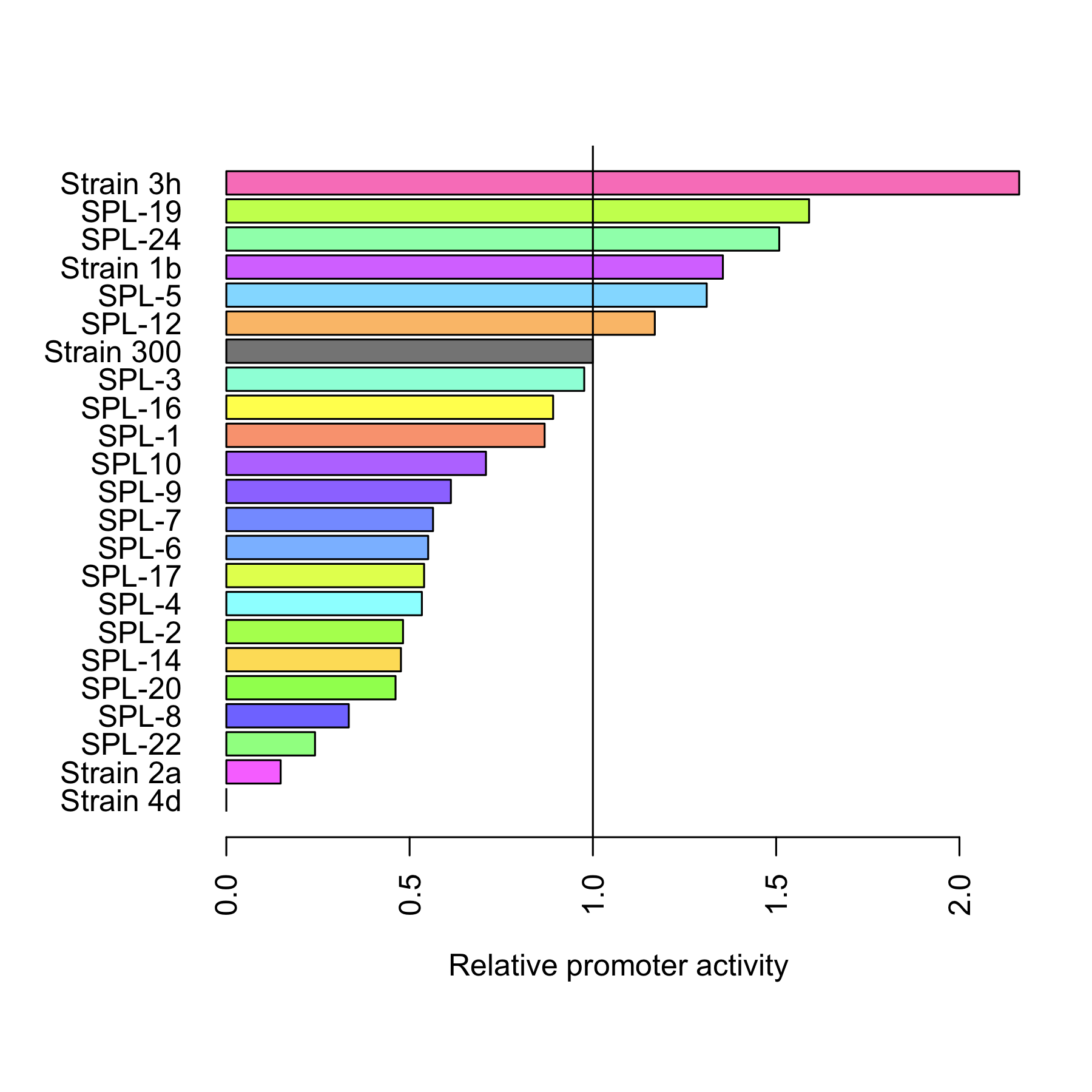
| - | [[File:DTU-Relative_promoter_activity.png|200px|right]] | + | [[File:DTU-Relative_promoter_activity.png|200px|right|link=Team:DTU-Denmark/Project_improving_araBAD]] |
| - | The dynamic range of the araBAD promoter was expanded. | + | The dynamic range of the araBAD promoter was expanded by either changing the -10 and -35 elements or randomly changing the promoter using a synthetic promoter library. Experiments were performed characterizing the constructed promoters. |
| - | + | ||
[[Team:DTU-Denmark/Project_improving_araBAD|Read more..]] | [[Team:DTU-Denmark/Project_improving_araBAD|Read more..]] | ||
| + | </div> | ||
| + | <div style="clear: both;"></div> | ||
| + | <div class="overviewBox right"> | ||
| + | |||
| + | == Data page == | ||
| + | [[File:DTU1_Data_page-figure1.png|200px|right|link=Team:DTU-Denmark/Data]] | ||
| + | The data page provides a description of the constructed BioBricks and how they work. It furthermore provides full links to the iGEM parts registry enabling easy retrieval of each submitted part. | ||
| + | [[Team:DTU-Denmark/Data|Read more...]] | ||
</div> | </div> | ||
| Line 71: | Line 72: | ||
== Results == | == Results == | ||
| - | + | The '''bioinformatic study''' revealed some interesting constrains for engineering novel sRNAs with our gene silencing tool; a terminal poly-U tail, a putative Hfq binding site, a stemloop without sequence constrains, and a terminal stemloop with high sequence conservation. See the results [[Team:DTU-Denmark/Bioinformatic#Results|here]]. | |
| - | + | '''Testing sRNA''' we were able to show that the target gene chiP can be placed on a plasmid and be regulated by a small RNA. Changing the complementary sequence removes this regulation. See the results [[Team:DTU-Denmark/Project_testing_sRNA#Results_and_Conclusions|here]]. | |
| - | + | Through rational design of the '''araBAD promoter''' we managed to reach a higher level of expression from that promoter. Also, we proved that Synthetic Promoter Library can be created for an inducible promoter to expand its dynamic range. See the results [[Team:DTU-Denmark/Project_improving_araBAD#Results|here]]. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | The '''modeling''' provides a guideline for [[Team:DTU-Denmark/Modeling#Parameters_and_proposed_experiments|determining parameters]] and design of [[Team:DTU-Denmark/Modeling#Design_of_dynamic_range|dynamic range]] of our gene silencing tool. A simulation revealed fast dynamics of gene silencing. See the results [[Team:DTU-Denmark/Modeling#Dynamics|here]]. | ||
</div> | </div> | ||
| - | |||
<div style="clear: both;"></div> | <div style="clear: both;"></div> | ||
Latest revision as of 03:41, 22 September 2011
Overview
Abstract
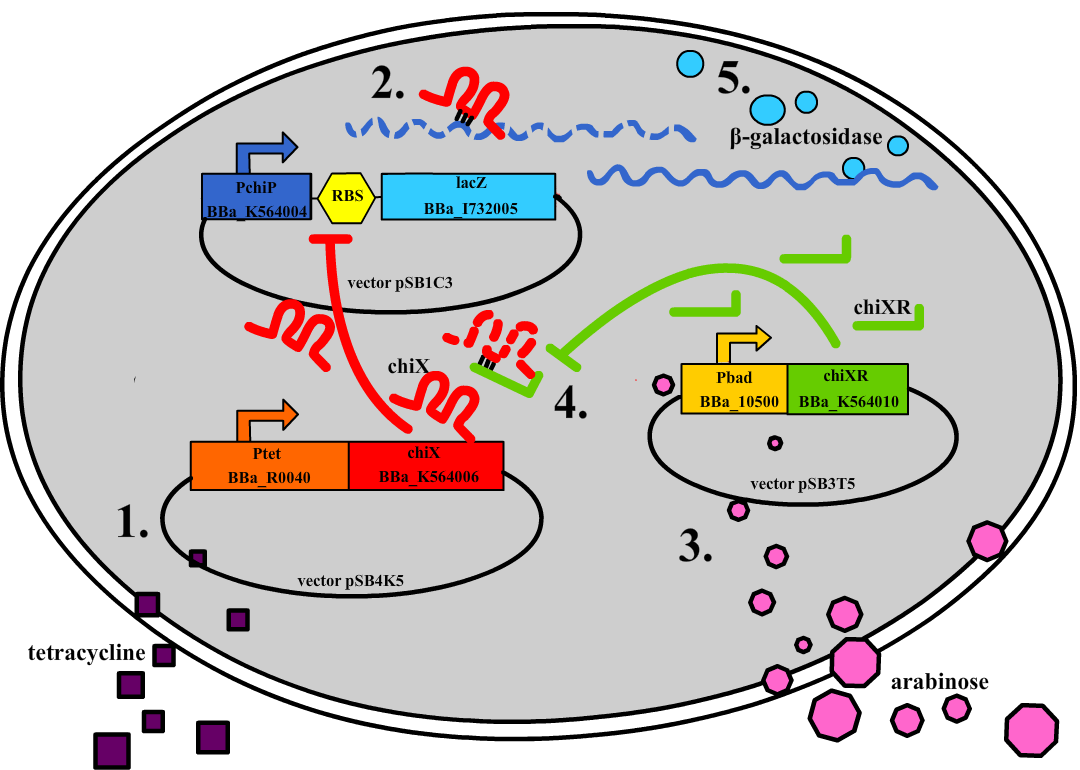
Small regulatory RNA is an active area of research with untapped possibilities for application in biotechnology. Such applications include convenient gene silencing and fine-tuning of gene expression, which are currently cumbersome processes restricted to well studied bacteria. We have investigated a novel type of RNA regulation based on the chitobiose system, where the inhibition caused by a small RNA is relieved by another small RNA called trap-RNA. We explore the possibility of using the system to uniquely target and repress any gene of interest, potentially providing unprecedented specificity and control of gene silencing. We furthermore constructed araBAD promoters with varying promoter activities using synthetic promoter libraries.Experiment: Testing sRNA
Experiments were performed to verify that the envisioned small RNA based gene silencing is possible. Plasmids containing and strains deleted for the components were constructed providing a biological model. Read more...
Bioinformatics
A bioinformatics study was performed to investigate the possibilities of engineering the trap-RNA system to target any gene. The study elucidates interesting features of sequence and secondary structure conservation guiding future application. Read more...
Modeling
A framework for characterization of gene silencing was developed to guide rational design and test hypotheses. Steady state analysis revealed that each trap-RNA system has a characteristic fold repression. Read more...
Experiment: Improving araBAD
The dynamic range of the araBAD promoter was expanded by either changing the -10 and -35 elements or randomly changing the promoter using a synthetic promoter library. Experiments were performed characterizing the constructed promoters. Read more..
Data page
The data page provides a description of the constructed BioBricks and how they work. It furthermore provides full links to the iGEM parts registry enabling easy retrieval of each submitted part. Read more...
Results
The bioinformatic study revealed some interesting constrains for engineering novel sRNAs with our gene silencing tool; a terminal poly-U tail, a putative Hfq binding site, a stemloop without sequence constrains, and a terminal stemloop with high sequence conservation. See the results here.
Testing sRNA we were able to show that the target gene chiP can be placed on a plasmid and be regulated by a small RNA. Changing the complementary sequence removes this regulation. See the results here.
Through rational design of the araBAD promoter we managed to reach a higher level of expression from that promoter. Also, we proved that Synthetic Promoter Library can be created for an inducible promoter to expand its dynamic range. See the results here.
The modeling provides a guideline for determining parameters and design of dynamic range of our gene silencing tool. A simulation revealed fast dynamics of gene silencing. See the results here.
 "
"