|
|
| (15 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | {{main}} | | {{main}} |
| | + | |
| | + | |
| | | | |
| | <html> | | <html> |
| - |
| + | <h2 class="art-postheader"> Results </h2> |
| - | <h2 class="art-postheader">Modelling</h2> | + | |
| | <div class="cleared"></div> | | <div class="cleared"></div> |
| - | <div class="art-postcontent"> | + | <div class="art-postcontent"> |
| - | | + | |
| - | <p><a name="indice"/> </p> | + | <!--------------------------------> |
| - | <table id="toc" class="toc"><tr><td><div id="toctitle"><h2>Contents</h2></div> | + | <!--------------------------------> |
| - | <ul> | + | <!--------------------------------> |
| - | <li class="toclevel-1"><a href="#Mathematical_modelling_page"><span class="tocnumber"></span> <span class="toctext">Mathematical modelling: introduction</span></a> | + | <!-----------MENU-----------------> |
| - | <ul> <br> | + | <!--------------------------------> |
| - | <li class="toclevel-2"><a href="#The importance of the mathematical model"><span class="tocnumber">1</span> <span class="toctext">The importance of mathematical modelling</span></a></li> | + | <!--------------------------------> |
| - | <li class="toclevel-2"><a href="#Equations_for_gene_networks"><span class="tocnumber">2</span> <span class="toctext">Equations for gene networks</span></a></li> | + | <!--------------------------------> |
| - | <ul>
| + | <br> |
| - | <li class="toclevel-3"><a href="#Hypothesis"><span class="tocnumber">2.1</span> <span class="toctext">Hypotheses</span></a></li> | + | <a name='indice'></a> |
| - | <li class="toclevel-3"><a href="#Equations_1_and_2"><span class="tocnumber">2.2</span> <span class="toctext">Equations (1) and (2)</span></a></li> | + | <div class="cleared"></div> |
| - | <li class="toclevel-3"><a href="#Equation_3"><span class="tocnumber">2.3</span> <span class="toctext">Equation (3)</span></a></li> | + | <div class="art-postcontent"> |
| - | <li class="toclevel-3"><a href="#Equation_4"><span class="tocnumber">2.4</span> <span class="toctext">Equation (4)</span></a></li> | + | <table id="toc" class="toc"> |
| - | </ul> | + | <tr> |
| | + | <td><div id="toctitle"> |
| | + | <h2>Contents</h2> |
| | + | </div> |
| | + | <ul> |
| | + | <li class="toclevel-1"><a href="#assembly"><span class="tocnumber">1</span> <span class="toctext">Part assembly</span></a> |
| | + | <li class="toclevel-1"><a href="#characterization"><span class="tocnumber">1</span> <span class="toctext">Characterization of basic modules</span></a> |
| | + | <ul> |
| | + | <li class="toclevel-2"><a href="#promoters"><span class="tocnumber">2.1</span> <span class="toctext">Promoter characterization</span></a></li> |
| | + | <li class="toclevel-2"><a href="#enzymes"><span class="tocnumber">2.2</span> <span class="toctext">Characterization of the activity of the enzymes AiiA and LuxI</span></a></li> |
| | + | <li class="toclevel-2"><a href="#rbs"><span class="tocnumber">2.3</span> <span class="toctext">Characterization of RBS efficiency</span></a></li> |
| | + | </ul> |
| | + | <li class="toclevel-1"><a href="#growth"><span class="tocnumber">3</span> <span class="toctext">Identification of bacterial growth parameters</span></a></li> |
| | + | <li class="toclevel-1"><a href="#HSL"><span class="tocnumber">4</span> <span class="toctext">Estimation of the spontaneous degradation of HSL in M9 medium and in cultures at different pH values</span></a></li> |
| | + | <li class="toclevel-1"><a href="#t9002"><span class="tocnumber">5</span> <span class="toctext">Characterization of BBa_T9002 biosensor</span></a></li> |
| | + | </ul></td> |
| | + | </tr> |
| | + | </table> |
| | + | </div> |
| | + | <script>if (window.showTocToggle) { var tocShowText = "show"; var tocHideText = "hide"; showTocToggle(); } </script> |
| | + | <em> NB: unless differently specified, all tests were performed in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Protocols#MG1655Z1'><em>E. coli</em> MGZ1</a> in M9 supplemented medium at 37°C. For the cloning of the parts, <a href='https://2011.igem.org/Team:UNIPV-Pavia/Protocols#TOP10'><em>E. coli</em> TOP10</a> was used. </em> <br> |
| | + | <br> |
| | + | |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!-----------ASSEMBLY-------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | |
| | + | <a name='assembly'></a> |
| | + | <h1>Parts assembly</h1> |
| | + | All the parts have been cloned with success. The part name, plasmids and quality controls are reported in the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Freezer'>Freezer section</a>. |
| | + | <div align="right"><small><a href="#indice" title="">^top</a></small></div> |
| | + | |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!-------CHARACTERIZATION---------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | |
| | + | <a name='characterization'></a> |
| | + | <h1>Characterization of basic modules</h1> |
| | + | |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------pTet and pLux-----------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | |
| | + | <a name='promoters'></a> |
| | + | <h2>Characterization of promoters pTet and pLux</h2> |
| | + | <p align='justify'> |
| | + | <p>Inducible and constitutive promoters were assembled upstream of different coding sequences containing an RBS from the Community collection.</p> |
| | + | <p>The assembled RBSs are:</p> |
| | + | <br> |
| | + | <div align='center'> |
| | + | <table class='data'> |
| | + | <tr> |
| | + | <td class="row"><b>BioBrick code</b></td> |
| | + | <td><b> Declared efficiency</b></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class="row">BBa_B0030 </td> |
| | + | <td class="row"> 0,6</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class="row">BBa_B0031 </td> |
| | + | <td class="row"> 0,07</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class="row">BBa_B0032 </td> |
| | + | <td class="row"> 0,3</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class="row">BBa_B0034 </td> |
| | + | <td class="row"> 1</td> |
| | + | </tr> |
| | + | </table> |
| | + | </div> |
| | + | <br> |
| | + | <div align="justify"> |
| | + | <p>For an inducible device, the RBS variation has the purpose to stretch the induction curve, thus modulating its PoPs-OUT range.</p> |
| | + | <p>The complex RBS-promoter acts as a whole regulatory element and determines the amount of translated protein. |
| | + | RBSs have been reported to have an un-modular behavior, since the translational efficiency is not independent on the coding sequences, but variates as an effect of different mRNA structure stability [Salis et al., Nat Biotec, 2009]. It is not possible to separate the effects of the sole promoter and of the sole RBS on the total amount/activity of gene product (in this case study, mRFP).</p> |
| | + | <p>For this reason, every combination 'Promoter+RBS' was studied as a different regulatory element. Regulatory elements were characterized using mRFP reporter protein for different RBSs in terms of Synthesis rate per Cell (<b>S<sub>cell</sub></b>) and <b>R.P.U.s</b> (Relative Promoter Units) as explained in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements'>measurements</a> section.</p> |
| | + | </p> |
| | + | <p>Operative parameters of the promoter are derived from the estimated Hill equations obtained by <em>nonlinear least squares</em> fitting (<em>lsqnonlin</em> Matlab routine) of the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Equations_for_gene_networks'>Hill function</a> expressed in RPUs:</p> |
| | + | <p></p> |
| | + | <p> |
| | + | <ol> |
| | + | <ul> |
| | + | <li><b> RPU<sub>max</sub></b> is equal to the α and represents the maximum promoter activity |
| | + | </p> |
| | + | </li> |
| | + | <p> |
| | + | <li><b> RPU<sub>min</sub></b> is equal to the α * δ represents the minimum promoter activity |
| | + | </p> |
| | + | </li> |
| | + | <p> |
| | + | <li> <b>Switch point</b> is computed as the abscissa of the inflection point of the Hill curve and it is representative of the position of linear region |
| | + | </p> |
| | + | </li> |
| | + | <p> |
| | + | <li> <b>Linearity boundaries</b> are determined as the intersection between the tangent line to the inflection point and the upper and lower horizontal boundaries of the Hill curve. |
| | | | |
| - | <li class="toclevel-2"><a href="#Table_of_parameters"><span class="tocnumber">3</span> <span class="toctext">Table of parameters</span></a></li>
| + | </li> |
| - | <ul>
| + | </p> |
| - | <li class="toclevel-3"><a href="#CV"><span class="tocnumber">3</span> <span class="toctext">Table of parameter CV</span></a></li>
| + | </ul> |
| - | </ul> | + | </ol> |
| - | | + | </p> |
| - | <li class="toclevel-2"><a href="#Parameter_estimation"><span class="tocnumber">4</span> <span class="toctext">Parameter estimation</span></a></li>
| + | <p align='justify'> The estimated parameters for the Hill functions of pLux are summarized in the table below. For more details on parameter estimation, see the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Ptet_&_Plux'>model section</a>. </p> |
| - | <ul>
| + | <table class='data' width='100%'> |
| - | <li class="toclevel-3"><a href="#Ptet_&_Plux"><span class="tocnumber">4.1</span> <span class="toctext">pTet & pLux</span></a></li>
| + | |
| - | <li class="toclevel-3"><a href="#introduction_to_T9002"><span class="tocnumber">4.2</span> <span class="toctext">T9002 introduction</span></a></li>
| + | |
| - | <li class="toclevel-3"><a href="#Enzymes"><span class="tocnumber">4.3</span> <span class="toctext"> AiiA & LuxI</span></a></li> | + | |
| - | <li class="toclevel-3"><a href="#N"><span class="tocnumber">4.4</span> <span class="toctext">N</span></a></li>
| + | |
| - | <li class="toclevel-3"><a href="#Degradation_rates"><span class="tocnumber">4.5</span> <span class="toctext">Degradation rates</span></a></li></ul>
| + | |
| - | | + | |
| - | | + | |
| - | <li class="toclevel-2"><a href="#Simulations"><span class="tocnumber">5</span> <span class="toctext">Simulations</span></a></li> <li class="toclevel-1"><a href="#Sensitivity_Analysis"><span class="tocnumber">6</span> <span class="toctext">Sensitivity Analysis of the steady state of enzyme expression in exponential phase</span></a></li>
| + | |
| - | <ul>
| + | |
| - | <li class="toclevel-2"><a href="#Steady state of enzyme expression"><span class="tocnumber">6.1</span> <span class="toctext">Steady state of enzyme expression</span></a></li>
| + | |
| - | <li class="toclevel-2"><a href="#Sensitivity analysis"><span class="tocnumber">6.2</span> <span class="toctext">Sensitivity analysis</span></a></li>
| + | |
| - | </ul>
| + | |
| - | <li class="toclevel-1"><a href="#References"><span class="tocnumber">7</span> <span class="toctext">References</span></a></li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | </ul>
| + | |
| - | </li>
| + | |
| - | </ul>
| + | |
| - | </td></tr></table>
| + | |
| - | <br><br>
| + | |
| - | <div class="listcircle">
| + | |
| - | | + | |
| - | <a name="Mathematical_modelling_page"></a><h1><span class="mw-headline"> <b>Mathematical modelling: introduction</b> </span></h1>
| + | |
| - | <div style='text-align:justify'><p>Mathematical modelling plays a central role in Synthetic Biology, due to its ability to serve as a crucial link between the concept and realization of a biological circuit: what we propose in this page is a mathematical modelling approach to the entire project, which has proven extremely useful before and after the "wet lab" activities.</p>
| + | |
| - | | + | |
| - | <p>Thus, immediately at the beginning, when there was little knowledge, a mathematical model based on a system of differential equations was derived and implemented using a set of reasonable values of model parameters, to validate the feasibility of the project. Once this became clear, starting from the characterization of each simple subpart created in the wet lab, some of the parameters of the mathematical model were estimated thanks to several ad-hoc experiments we performed within the iGEM project (others were derived from literature) and they were used to predict the final behaviour of the whole engineered closed-loop circuit. This approach is consistent with the typical one adopted for the analysis and synthesis of a biological circuit, as exemplified by <a href="#Pasotti"><i><b>Pasotti L</b> et al. 2011.</i></a></p>
| + | |
| - | | + | |
| - | <p>After a brief overview on the importance of the mathematical modelling approach, we deeply analyze the system of equations, underlining the role and function of the parameters involved.</p>
| + | |
| - | <p>Experimental procedures for parameter estimation are discussed and, finally, a different type of circuit is presented. <font color="red">Simulations were performed, using <em>ODEs</em> with MATLAB and used to explain the difference between a closed-loop control system model and an open one.</font></div></p>
| + | |
| - | <div align="right"><small><a href="#indice">^top</a></small></div>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | | + | |
| - | <a name="The importance of the mathematical model"></a><h2> <span class="mw-headline"> <b>The importance of mathematical modelling</b> </span></h2>
| + | |
| - | <div style='text-align:justify'><p>Mathematical modelling reveals fundamental in the challenge of understanding and engineering complex biological systems. Indeed, these are characterized by a high degree of interconnection among the single constituent parts, requiring a comprehensive analysis of their behavior through mathematical formalisms and computational tools.</p>
| + | |
| - | <div>Synthetically, we can identify two major roles concerning mathematical models:</div>
| + | |
| - | | + | |
| - | <ul>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <p><li><b>Simulation</b>: mathematical models allow to analyse complex system dynamics and to reveal the relationships between the involved variables, starting from the knowledge of the single subparts behavior and from simple hypotheses of their interconnection. <a href="#Endler">(<i><b>Endler L</b> et al. 2009</i>)</a></li></p>
| + | |
| - | | + | |
| - | <p><li><b>Knowledge elicitation</b>: mathematical models summarize into a small set of parameters the results of several experiments (parameter identification), allowing a robust comparison among different experimental conditions and providing an efficient way to synthesize knowledge about biological processes. Then, through the simulation process, they make possible the re-usability of the knowledge coming from different experiments, engineering complex systems from the composition of its constituent subparts under appropriate experimental/environmental conditions <a href="#Braun">(<i><b>Braun D</b> et al. 2005</a>;<a href="#Canton"> <b>Canton B</b> et al 2008</a></i>)</font>.</li>
| + | |
| - | </p>
| + | |
| - | | + | |
| - | </ul>
| + | |
| - | </div>
| + | |
| - | <div align="right"><small><a href="#indice">^top</a></small></div>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <a name="Equations_for_gene_networks"></a><h2> <span class="mw-headline"> <b>Equations for gene networks</b> </span></h2>
| + | |
| - | <p>Below is provided the system of equations of our mathematical model. </p>
| + | |
| - | | + | |
| - | <div style='text-align:justify'><div class="thumbinner" style="width: 850px;"><img alt="" src="https://static.igem.org/mediawiki/2011/4/43/Model_new.jpg" class="thumbimage" height="55%" width="80%"></a></div></div>
| + | |
| - | <br>
| + | |
| - | <table align='center' width='100%'>
| + | |
| - | <tr>
| + | |
| - | <td>
| + | |
| - | <div style='text-align:center'><div class="thumbinner" style="width:100%;"><img alt="" src="https://static.igem.org/mediawiki/2011/5/5e/Circuito_finale.jpg" class="thumbimage" width="87%"></a></div></div>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | <div style='text-align:justify'><div class="thumbinner" style="width: 850px;"><img alt="" src="https://static.igem.org/mediawiki/2011/c/c7/QS_system_synthetic_circuit.png" class="thumbimage" width="85%"></a></div></div>
| + | |
| - | <div align="right"><small><a href="#indice">^top</a></small></div>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <a name="Hypothesis"></a><h4> <span class="mw-headline"> <b>Hypotheses of the model</b> </span></h4>
| + | |
| - | <table class="data">
| + | |
| - | <tr>
| + | |
| - | <td>
| + | |
| - | <div style='text-align:justify'>
| + | |
| - | <em>
| + | |
| - | <b>HP<sub>1</sub></b>: in equation (2) only HSL is considered as inducer, instead of the complex LuxR-HSL.
| + | |
| - | This is motivated by the fact that our final device offers a constitutive LuxR production due to the upstream constitutive promoter Pλ. Assuming LuxR is abundant in the cytoplasm, we can understand this simplification of attributing pLux promoter induction only by HSL.
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <b>HP<sub>2</sub></b>: in system equation, LuxI and AiiA amounts are expressed per cell. For this reason, the whole equation (3), except for the
| + | |
| - | term of intrinsic degradation of HSL, is multiplied by the number of cells N, due to the property of the lactone to diffuse freely inside/outside bacteria.
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <b>HP<sub>3</sub></b>: as regards promoters pTet and pLux, we assume their strengths (measured in PoPs), due to a given concentration of inducer (aTc and HSL for Ptet and Plux respectively), to be
| + | |
| - | independent from the gene downstream.
| + | |
| - | In other words, in our hypothesis, if the mRFP coding region is substituted with a region coding for another gene (in our case, AiiA or LuxI), we would obtain the same synthesis rate:
| + | |
| - | this is the reason why the strength of the complex promoter-RBS is expressed in Arbitrary Units [AUr].
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | <b>HP<sub>4</sub></b>: considering the exponential growth, the enzymes AiiA and LuxI concentration is supposed to be constant, because their production is equally compensated by dilution.
| + | |
| - | </em>
| + | |
| - | </div>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | <br>
| + | |
| - | <div align="right"><small><a href="#indice">^top</a></small></div>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | <a name="Equations_1_and_2"></a><h4> <span class="mw-headline"> <b>Equations (1) and (2)</b> </span></h4>
| + | |
| - | <div style='text-align:justify'><p>Equations (1) and (2) have identical structure, differing only in the parameters involved. They represent the synthesis, degradation and diluition of both the enzymes in the circuit, LuxI and AiiA, respectively in the first and second equation: in each of them both transcription and translation processes have been condensed. The mathematical formulation is analogous to the one used by <a href="#Pasotti"><i><b>Pasotti L</b> et al. 2011</i></a>, Suppl. Inf., even if we do not take LuxR-HSL complex formation into account, as explained below.</p>
| + | |
| - | <p>These equations are composed of 2 parts:</p> | + | |
| - | <ol>
| + | |
| - | <li>The first term describes, through Hill's equation, the synthesis rate of the protein of interest (either LuxI or AiiA) depending on the concentration of the inducer (anhydrotetracicline -aTc- or HSL respectively), responsible for the activation of the regulatory element composed of promoter and RBS. In the parameter table (see below), α refers to the maximum activation of the promoter, while δ stands for its leakage activity (this means that the promoter is slightly active even if there is no induction). In particular, in equation (1), the almost entire inhibition of pTet promoter is given by the constitutive production of TetR by our MGZ1 strain. In equation (2), pLux is almost inactive in the absence of the complex LuxR-HSL. Furthermore, in both equations k stands for the dissociation constant of the promoter from the inducer (respectively aTc and HSL in eq. 1 and 2), while η is the cooperativity constant.</p>
| + | |
| - | <p><li>The second term in equations (1) and (2) is in turn composed of 2 parts. The former one (<em>γ</em>*LuxI or <em>γ</em>*AiiA respectively) describes, with an exponential decay, the degradation rate per cell of the protein. The latter (μ*(Nmax-N)/Nmax)*LuxI or μ*(Nmax-N)/Nmax)*AiiA, respectively) takes into account the dilution factor against cell growth which is related to the cell replication process.</p>
| + | |
| - | </ol>
| + | |
| - | </div>
| + | |
| - | <div align="right"><small><a href="#indice">^top</a></small></div>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <a name="Equation_3"></a><h4> <span class="mw-headline"> <b>Equation (3)</b> </span></h4>
| + | |
| - | <div style='text-align:justify'><p>Here the kinetics of HSL is modeled, through enzymatic reactions either related to the production or the degradation of HSL. This equation is composed of 3 parts: </p>
| + | |
| - | <ol>
| + | |
| - | <p><li> The first term represents the production of HSL due to LuxI expression. We modeled this process with a saturation curve in which V<sub>max</sub> is the HSL maximum transcription rate, while k<sub>M,LuxI</sub> is LuxI dependent half-saturation constant.</p>
| + | |
| - | <p><li> The second term represents the degradation of HSL due to the AiiA expression. Similarly to LuxI, k<sub>cat</sub> represents the maximum degradation per unit of HSL concentration, while k<sub>M,AiiA</sub> is the concentration at which AiiA dependent HSL degradation rate is (k<sub>cat</sub>*HSL)/2. The formalism is similar to that found in the Supplementary Information of <a href="#Danino"><i><b>Danino T</b> et al 2010.</i></a></font></p>
| + | |
| - | <p><li> The third term (γ<sub>HSL</sub>*HSL) is similar to the corresponding ones present in the first two equations and describes the intrinsic protein degradation.</div>
| + | |
| - | <div align="right"><small><a href="#indice">^top</a></small></div></p>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | | + | |
| - | <a name="Equation_4"></a><h4> <span class="mw-headline"> <b>Equation (4)</b> </span></h4>
| + | |
| - | <div style='text-align:justify'>This is the typical cells growth equation, depending on the rate μ and the maximum number N<sub>max</sub> of cells per well reachable <a href="#Pasotti">(<i><b>Pasotti L</b> et al. 2009</i>).</a></div>
| + | |
| - | <div align="right"><br><small><a href="#indice">^top</a></small></div>
| + | |
| - | <br><br>
| + | |
| - | | + | |
| - | | + | |
| - | <a name="Table_of_parameters"></a><h2> <span class="mw-headline"> <b>Table of parameters and species</b> </span></h2>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | <center>
| + | |
| - | <table class="data">
| + | |
| | <tr> | | <tr> |
| - | <td class="row"><b>Parameter & Species</b></td>
| + | <td class="row"><b>RBS</b></td> |
| - | <td class="row"><b>Description</b></td>
| + | <td class="row"><b>α<sub>p<sub>Lux</sub></sub> [(AUr/min)/cell]</b></td> |
| - | <td class="row"><b>Unit of Measurement</b></td>
| + | <td class="row"><b>δ<sub>p<sub>Lux</sub></sub> [-]</b></td> |
| - | <td class="row"><b>Value</b></td>
| + | <td class="row"><b>η<sub>p<sub>Lux</sub></sub> [-]</b></td> |
| - | </tr>
| + | <td class="row"><b>k<sub>p<sub>Lux</sub></sub> [ng/ml]</b></td> |
| - | | + | </tr> |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">α<sub>p<sub>Tet</sub></sub></td>
| + | |
| - | <td class="row">maximum transcription rate of pTet (dependent on <a href="#RBS">RBSx</a> efficiency)</td>
| + | |
| - | <td class="row">[(AUr/min)/cell]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">δ<sub>p<sub>Tet</sub></sub></td> | + | |
| - | <td class="row">leakage factor of promoter pTet basic activity</td>
| + | |
| - | <td class="row">[-]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">η<sub>p<sub>Tet</sub></sub></td>
| + | |
| - | <td class="row">Hill coefficient of pTet</td>
| + | |
| - | <td class="row">[-]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">k<sub>p<sub>Tet</sub></sub></td> | + | |
| - | <td class="row">dissociation constant of aTc from pTet</td>
| + | |
| - | <td class="row">[ng/ml]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">α<sub>p<sub>Lux</sub></sub></td>
| + | |
| - | <td class="row">maximum transcription rate of pLux (dependent on <a href="#RBS">RBSx</a> efficiency)</td>
| + | |
| - | <td class="row">[(AUr/min)/cell]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">δ<sub>p<sub>Lux</sub></sub></td>
| + | |
| - | <td class="row">leakage factor of promoter pLux basic activity</td>
| + | |
| - | <td class="row">[-]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">η<sub>p<sub>Lux</sub></sub></td> | + | |
| - | <td class="row">Hill coefficient of pLux</td>
| + | |
| - | <td class="row">[-]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">k<sub>p<sub>Lux</sub></sub></td>
| + | |
| - | <td class="row">dissociation constant of HSL from pLux</td>
| + | |
| - | <td class="row">[nM]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">γ<sub>p<sub>Lux</sub></sub></td>
| + | |
| - | <td class="row">LuxI constant degradation</td>
| + | |
| - | <td class="row">[1/min]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td class="row">γ<sub>AiiA</sub></td>
| + | |
| - | <td class="row">AiiA constant degradation</td>
| + | |
| - | <td class="row">[1/min]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">γ<sub>HSL</sub></td>
| + | |
| - | <td class="row">HSL constant degradation</td>
| + | |
| - | <td class="row">[1/min]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">V<sub>max</sub></td>
| + | |
| - | <td class="row">maximum transcription rate of LuxI</td>
| + | |
| - | <td class="row">[nM/(min*cell)]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">k<sub>M,LuxI</sub></td>
| + | |
| - | <td class="row">half-saturation constant of LuxI from HSL</td>
| + | |
| - | <td class="row">[AUr/cell]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td class="row">k<sub>cat</sub></td>
| + | |
| - | <td class="row">maximum number of enzymatic reactions catalyzed per minute</td>
| + | |
| - | <td class="row">[1/(min*cell)]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">k<sub>M,AiiA</sub></td>
| + | |
| - | <td class="row">half-saturation constant of AiiA from HSL</td>
| + | |
| - | <td class="row">[AUr/cell]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">N<sub>max</sub></td>
| + | |
| - | <td class="row">maximum number of bacteria per well</td>
| + | |
| - | <td class="row">[cell]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | <tr>
| + | |
| - | <td class="row">μ</td>
| + | |
| - | <td class="row">rate of bacteria growth</td>
| + | |
| - | <td class="row">[1/min]</td>
| + | |
| - | <td class="row">-</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| - | | + | |
| | <tr> | | <tr> |
| - | <td class="row">LuxI</td>
| + | <td class="row">BBa_B0030</td> |
| - | <td class="row">kinetics of LuxI enzyme</td>
| + | <td class="row">438 [10]</td> |
| - | <td class="row">[<sup>AUr</sup>⁄<sub>cell</sub>]</td>
| + | <td class="row">0.05 [>100]</td> |
| - | <td class="row">-</td>
| + | <td class="row">2 [47]</td> |
| - | </tr>
| + | <td class="row">1.88 [27]</td> |
| - | | + | </tr> |
| | <tr> | | <tr> |
| - | <td class="row">AiiA</td>
| + | <td class="row">BBa_B0031</td> |
| - | <td class="row">kinetics of AiiA enzyme</td>
| + | <td class="row">9.8 [7]</td> |
| - | <td class="row">[<sup>AUr</sup>⁄<sub>cell</sub>]</td>
| + | <td class="row">0.11 [57]</td> |
| - | <td class="row">-</td>
| + | <td class="row">1.2 [29]</td> |
| - | </tr>
| + | <td class="row">1.5 [26]</td> |
| - | | + | </tr> |
| | <tr> | | <tr> |
| - | <td class="row">HSL</td>
| + | <td class="row">BBa_B0032</td> |
| - | <td class="row">kinetics of HSL</b></td>
| + | <td class="row">206 [3]</td> |
| - | <td class="row">[<sup>nM</sup>⁄<sub>(min)</sub>]</td>
| + | <td class="row">0 [>>100]</td> |
| - | <td class="row">-</td>
| + | <td class="row">1.36 [10]</td> |
| - | </tr>
| + | <td class="row">1.87 [9]</td> |
| - | | + | </tr> |
| | <tr> | | <tr> |
| - | <td class="row">N</td>
| + | <td class="row">BBa_B0034</td> |
| - | <td class="row">number of cells</td>
| + | <td class="row">1105 [6]</td> |
| - | <td class="row">cell</td>
| + | <td class="row">0.02 [>100]</td> |
| - | <td class="row">-</td>
| + | <td class="row">1.33 [19]</td> |
| - | </tr>
| + | <td class="row">2.34 [18]</td> |
| | + | </tr> |
| | + | </table> |
| | + | <div align="center">Data are provided as average [CV%].</div> |
| | + | <br> |
| | + | <p>The operative parameters are summarized in the table below:</p> |
| | + | </div> |
| | + | <table align='center' class='data' width='100%'> |
| | + | <tr> |
| | + | <td class='row'><b>RBS</b></td> |
| | + | <td class='row'><b>RPU<sub>max</sub></b></td> |
| | + | <td class='row'><b>RPU<sub>min</sub></b></td> |
| | + | <td class='row'><b>Switch point [nM]</b></td> |
| | + | <td class='row'><b>Linear boundaries [MIN; MAX] [nM]</b></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0030</td> |
| | + | <td class='row'>4.28</td> |
| | + | <td class='row'>0.20</td> |
| | + | <td class='row'>1.08</td> |
| | + | <td class='row'>[0.36; 3.27]</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0031</td> |
| | + | <td class='row'>4.93</td> |
| | + | <td class='row'>0.55</td> |
| | + | <td class='row'>0.25</td> |
| | + | <td class='row'>[0.03; 2.30]</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0032</td> |
| | + | <td class='row'>9.49</td> |
| | + | <td class='row'>0.02</td> |
| | + | <td class='row'>0.47</td> |
| | + | <td class='row'>[0.07; 3.07]</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0034</td> |
| | + | <td class='row'>21.53</td> |
| | + | <td class='row'>0.51</td> |
| | + | <td class='row'>0.53</td> |
| | + | <td class='row'>[0.08; 3.77]</td> |
| | + | </tr> |
| | </table> | | </table> |
| - | </center> | + | <table width='100%'> |
| - | <br> | + | <tr> |
| - | <br> | + | <td width='50%'><div style="text-align:justify"> |
| | + | <div class="thumbinner" width='100%'><a href="https://static.igem.org/mediawiki/2011/5/50/E17_RPU_80.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/5/50/E17_RPU_80.jpg" class="thumbimage" width="100%"></a></div> |
| | + | </div></td> |
| | + | <td width='50%'><div style="text-align:justify"> |
| | + | <div class="thumbinner" width='100%'><a href="https://static.igem.org/mediawiki/2011/c/c4/E18_RPU_80.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/c/c4/E18_RPU_80.jpg" class="thumbimage" width="100%"></a></div> |
| | + | </div></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td width='50%'><div style="text-align:justify"> |
| | + | <div class="thumbinner" width='100%'><a href="https://static.igem.org/mediawiki/2011/7/78/E19_RPU_80.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/7/78/E19_RPU_80.jpg" class="thumbimage" width="100%"></a></div> |
| | + | </div></td> |
| | + | <td width='50%'><div style="text-align:justify"> |
| | + | <div class="thumbinner" width='100%'><a href="https://static.igem.org/mediawiki/2011/5/55/E20_RPU_80.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/5/55/E20_RPU_80.jpg" class="thumbimage" width="100%"></a></div> |
| | + | </div></td> |
| | + | </tr> |
| | + | </table> |
| | + | <p>The protocols for the characterization of p<sub>Tet</sub> promoter are reported in the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#pTet_protocol'>p<sub>Tet</sub> measurement section</a>.</p> |
| | + | <p> Here we have characterized its transcriptional strength as a function of aTc induction (ng/ul) for different RBSs. Three different induction curves were obtained and are reported in figure:</p> |
| | | | |
| - |
| |
| - | <div align="justify"><b><a name="RBS">NOTE</a></b><p>In order to better investigate the range of dynamics of each subpart, every promoter has been studied with 4 different RBSs, so as to develop more knowledge about the state variables in several configurations of RBS' efficiency <a href="#Salis">(<i><b>Salis HM</b> et al. 2009</i>)</a>. Hereafter, referring to the notation "RBSx" we mean, respectively,
| |
| - | <a href="http://partsregistry.org/Part:BBa_B0030">RBS30</a>,
| |
| - | <a href="http://partsregistry.org/Part:BBa_B0031">RBS31</a>,
| |
| - | <a href="http://partsregistry.org/Part:BBa_B0032">RBS32</a>,
| |
| - | <a href="http://partsregistry.org/Part:BBa_B0034">RBS34</a>.
| |
| - | </p></div>
| |
| - | <br>
| |
| - | <div align="right"><small><a href="#indice">^top</a></small></div>
| |
| - |
| |
| - | <a name="CV"></a><h4> <span class="mw-headline"> <b>Parameter CV</b> </span></h4>
| |
| | <center> | | <center> |
| - | <table class="data"> | + | <a href="https://static.igem.org/mediawiki/2011/a/af/E32_RPU_80.jpg" class="image"> <img alt="" src="https://static.igem.org/mediawiki/2011/a/af/E32_RPU_80.jpg" class="thumbimage" width="50%"></a> |
| - | <tr>
| + | </center> |
| - | <td class="row"><b>Parameter & Species</b></td>
| + | <table width='100%'> |
| - | <td class="row"><b>BBa_B0030</b></td>
| + | <tr> |
| - | <td class="row"><b>BBa_B0031</b></td>
| + | <td width='50%'><div style="text-align:justify"> |
| - | <td class="row"><b>BBa_B0032</b></td>
| + | <div class="thumbinner" width='100%'><a href="https://static.igem.org/mediawiki/2011/9/99/E34_RPU_80.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/9/99/E34_RPU_80.jpg" class="thumbimage" width="100%"></a></div> |
| - | <td class="row"><b>BBa_B0034</b></td>
| + | </div></td> |
| - | </tr>
| + | <td width='50%'><div style="text-align:justify"> |
| - | <tr>
| + | <div class="thumbinner" width='100%'><a href="https://static.igem.org/mediawiki/2011/e/e4/E35_RPU_80.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/e/e4/E35_RPU_80.jpg" class="thumbimage" width="100%"></a></div> |
| - | <td class="row">α<sub>p<sub>Tet</sub></sub></td>
| + | </div></td> |
| - | <td class="row">3.7</td>
| + | </tr> |
| - | <td class="row">ND</td>
| + | |
| - | <td class="row">12</td>
| + | |
| - | <td class="row">5.94</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td class="row">δ<sub>p<sub>Tet</sub></sub></td>
| + | |
| - | <td class="row">91.61</td>
| + | |
| - | <td class="row">>>100</td>
| + | |
| - | <td class="row">>100</td>
| + | |
| - | <td class="row">40.59</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td class="row">η<sub>p<sub>Tet</sub></sub></td>
| + | |
| - | <td class="row">23.72</td>
| + | |
| - | <td class="row">>>100</td>
| + | |
| - | <td class="row">57.62</td>
| + | |
| - | <td class="row">47.6</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td class="row">k<sub>p<sub>Tet</sub></sub></td>
| + | |
| - | <td class="row">4.16</td>
| + | |
| - | <td class="row">>>100</td>
| + | |
| - | <td class="row">14.99</td>
| + | |
| - | <td class="row">5.43</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td class="row">α<sub>p<sub>Lux</sub></sub></td>
| + | |
| - | <td class="row">10.14</td>
| + | |
| - | <td class="row">7.13</td>
| + | |
| - | <td class="row">2.78</td>
| + | |
| - | <td class="row">5.8</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td class="row">δ<sub>p<sub>Lux</sub></sub></td>
| + | |
| - | <td class="row">179.7</td>
| + | |
| - | <td class="row">57.04</td>
| + | |
| - | <td class="row">1317.7</td>
| + | |
| - | <td class="row">187.2</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td class="row">η<sub>p<sub>Lux</sub></sub></td>
| + | |
| - | <td class="row">47.73</td>
| + | |
| - | <td class="row">29.13</td>
| + | |
| - | <td class="row">9.75</td>
| + | |
| - | <td class="row">19.3</td>
| + | |
| - | </tr>
| + | |
| - | <tr>
| + | |
| - | <td class="row">k<sub>p<sub>Lux</sub></sub></td>
| + | |
| - | <td class="row">27.5</td>
| + | |
| - | <td class="row">25.81</td>
| + | |
| - | <td class="row">8.46</td>
| + | |
| - | <td class="row">17.86</td>
| + | |
| - | </tr>
| + | |
| - | | + | |
| | </table> | | </table> |
| - | </center> | + | <p align='justify'> The estimated parameters of the Hill curves described in the figures are summarized in the table below: </p> |
| - | <div align="right"><br><small><a href="#indice">^top</a></small></div>
| + | |
| | <br> | | <br> |
| - | <br> | + | <table class='data' width='100%' title='parameter value'> |
| - | | + | <tr> |
| - | | + | <td class="row"><b>RBS</b></td> |
| - | | + | <td class="row"><b>α<sub>p<sub>Tet</sub></sub> [(AUr/min)/cell]</b></td> |
| - | | + | <td class="row"><b>δ<sub>p<sub>Tet</sub></sub> [-]</b></td> |
| - | <a name="Parameter_estimation"></a><h2> <span class="mw-headline"> <b>Parameter estimation</b></span></h2> | + | <td class="row"><b>η<sub>p<sub>Tet</sub></sub> [-]</b></td> |
| - | <div style='text-align:justify'>The aim of the model is to predict the behavior of the final closed loop circuit starting from the characterization of single BioBrick parts through a set of well-designed <em>ad hoc</em> experiments. This section presents the experiments performed. | + | <td class="row"><b>k<sub>p<sub>Tet</sub></sub> [nM]</b></td> |
| - | As explained before in <a href="#RBS"><span class="toctext"><b>NOTE</b></span></a>, considering a set of 4 RBSs for each subpart expands the range of dynamics and helps us to better understand the interactions between state variables.
| + | </tr> |
| - | </div> | + | <tr> |
| - | <br><div align="right"><small><a href="#indice">^top</a></small></div> | + | <td class="row">BBa_B0030</td> |
| - | <br> | + | <td class="row">230.67 [3.7]</td> |
| - | | + | <td class="row">0.028 [91.61]</td> |
| - | | + | <td class="row">4.61 [23.73]</td> |
| - | | + | <td class="row">8.75 [4.16]</td> |
| - | <a name="Ptet_&_Plux"></a><h4> <span class="mw-headline"> <b>Promoter (PTet & pLux)</b> </span></h4> | + | </tr> |
| - | <div style='text-align:justify'> | + | <tr> |
| - | | + | <td class="row">BBa_B0031</td> |
| - | <table align='center' width='100%'> | + | <td class="row">ND</td> |
| - | <tr> | + | <td class="row">ND</td> |
| - | <td> | + | <td class="row">ND</td> |
| - | <div style='text-align:center'><div class="thumbinner" style="width:100%;"><img alt="" src="https://static.igem.org/mediawiki/2011/9/91/Caratterizzazione_ptetN.jpg" class="thumbimage" width="33%"></a></div></div> | + | <td class="row">ND</td> |
| - | </td> | + | </tr> |
| - | </tr> | + | <tr> |
| | + | <td class="row">BBa_B0032</td> |
| | + | <td class="row">55.77 [12]</td> |
| | + | <td class="row">1.53E-11 [>>100]</td> |
| | + | <td class="row">4.98 [57.62]</td> |
| | + | <td class="row">7.26 [14.98]</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class="row">BBa_B0034</td> |
| | + | <td class="row">120 [5.95]</td> |
| | + | <td class="row">0.085 [40.6]</td> |
| | + | <td class="row">24.85 [47.6]</td> |
| | + | <td class="row">9 [5.43]</td> |
| | + | </tr> |
| | </table> | | </table> |
| - | | + | Data are provided as average [CV%] <br> |
| - | | + | |
| - | </div>
| + | |
| - | | + | |
| - | <div style='text-align:justify'>
| + | |
| - | | + | |
| - | <table align='center' width='100%'>
| + | |
| - | <tr>
| + | |
| - | <td>
| + | |
| - | <div style='text-align:center'><div class="thumbinner" style="width:100%;"><img alt="" src="https://static.igem.org/mediawiki/2011/7/79/Caratterizzazione_pluxN.jpg" class="thumbimage" width="70%"></a></div></div>
| + | |
| - | </td>
| + | |
| - | </tr>
| + | |
| - | </table>
| + | |
| - | | + | |
| - | | + | |
| - | </div>
| + | |
| - | <div style='text-align:justify'><p>These are the first parts tested, with the target of learning more about pTet and pLux promoters. In particular, as previously explained in <a href=#RBS>NOTE</a>, for each promoter, we tested four different combinations of promoter-RBS, providing us a set of fundamental building blocks for the subsequent assebly of the closed-loop circuit.</p>
| + | |
| - | <p>As shown in the figure below, we considered a range of inductions and we monitored, in time, absorbance (O.D. stands for "optical density") and fluorescence; the two vertical segments for each graph highlight the exponential phase of bacterial growth. S<sub>cell</sub> (namely, synthesis rate per cell) can be derived as a function of inducer concentration, thereby providing the desired input-output relation (inducer concentration versus promoter+RBS activity), which was modelled as a Hill curve:</p>
| + | |
| - | | + | |
| - | <div align="center"><div class="thumbinner" style="width: 600px;"><img alt="" src="https://static.igem.org/mediawiki/2011/5/58/Scell.jpg" class="thumbimage" width="45%"></a></div></div>
| + | |
| - | | + | |
| - | However, also Relative Promoter Unit (RPU, <a href="#Kelly"><i><b>Kelly JR</b> et al. 2009</i></a>) has been calculated as a ratio of S<sub>cell</sub> of promoter of interest and the S<sub>cell</sub> of <a href="http://partsregistry.org/Part:BBa_J23101">BBa_J23101</a> (reference to <a href="#Hypothesis"><span class="toctext"><b><em>HP<sub>3</sub></em></b></span></a>).<br>
| + | |
| - | | + | |
| - | <div style='text-align:center'><div class="thumbinner" style="width: 600px;"><img alt="" src="https://static.igem.org/mediawiki/2011/2/26/Box2_new.jpg" class="thumbimage" height="48%" width="120%"></a></div></div><br>
| + | |
| - | | + | |
| - | <p>As shown in the figure, α, as already mentioned, represents the protein maximum synthesis rate, which is reached, in accordance with Hill equation, when the inducer concentration tends to infinite, and, more practically, when the inducer concentration is sufficiently higher than the dissociation constant. Meanwhile the product α*δ stands for the leakage activity (at no induction), liable for protein production (LuxI and AiiA respectively) even in the absence of inducer.</p>
| + | |
| - | <p>The paramenter η is the Hill's cooperativity constant and it affects the ripidity of transition from the lower and upper boundary of the curve relating S<sub>cell</sub> to the inducer concentration.
| + | |
| - | Lastly, k stands for the semi-saturation constant and, in case of η=1, it indicates the concentration of substrate at which half the synthesis rate is achieved.
| + | |
| - | <br><div align="right"><small><a href="#indice">^top</a></small></div>
| + | |
| | <br> | | <br> |
| - | <br> | + | <p align='justify'> The operative parameters are summarized in the table below: </p> |
| - | | + | <table align='center' class='data' width='100%'> |
| - | | + | <tr> |
| - | | + | <td class='row'><b>RBS</b></td> |
| - | <a name="introduction_to_T9002"></a><h4> <span class="mw-headline"> <b>T9002 introduction</b> </span></h4>
| + | <td class='row'><b>RPU<sub>max</sub></b></td> |
| - | <div style='text-align:justify'> | + | <td class='row'><b>RPU<sub>min</sub></b></td> |
| - | <em> | + | <td class='row'><b>Switch point [ng/ml]</b></td> |
| - | <p>LuxI and AiiA tests have been always performed exploiting the well-characterized BioBrick <a href="http://partsregistry.org/Part:BBa_T9002">BBa_T9002</a>, by which it's possible to quantify exactly the concentration of HSL.</p> | + | <td class='row'><b>Linear boundaries [MIN; MAX] [ng/ml]</b></td> |
| - | <div align="center"><div class="thumbinner" style="width: 500px;"><img alt="" src="https://static.igem.org/mediawiki/2011/c/c2/T9002.jpg" class="thumbimage" width="110%"></a></div></div> | + | </tr> |
| - | <p>This is a biosensor which receives HSL concentration as input and returns GFP intensity (more precisely S<sub>cell</sub>) as output.<a href="#Canton"> (<i><b>Canton</b> et al. 2008</i>).</a> | + | <tr> |
| - | According to this, it is necessary to understand the input-output relationship: so, a T9002 "calibration" curve is plotted for each test performed.</p><br><br>
| + | <td class='row'>B0030</td> |
| - | </em> | + | <td class='row'>1.53</td> |
| - | </div> | + | <td class='row'>~0</td> |
| - | <div align="right"><small><a href="#indice">^top</a></small></div> | + | <td class='row'>7.95</td> |
| - | <br> | + | <td class='row'>[4.66;11.99]</td> |
| - | | + | </tr> |
| - | | + | <tr> |
| - | | + | <td class='row'>B0031</td> |
| - | <a name="Enzymes"></a><h4> <span class="mw-headline"> <b>AiiA & LuxI</b> </span></h4> | + | <td class='row'>ND</td> |
| - | <div style='text-align:justify'><p>This paragraph explains how parameters of equation (3) are estimated. The target is to learn the AiiA degradation and LuxI production mechanisms in addiction to HSL intrinsic degradation, in order to estimate V<sub>max</sub>, K<sub>M,LuxI</sub>, k<sub>cat</sub>, K<sub>M,AiiA</sub> and γ<sub>HSL</sub> parameters. We adopt tests composed of two steps. In the first one, the following BioBrick parts are used:</p> | + | <td class='row'>ND</td> |
| - | </div> | + | <td class='row'>ND</td> |
| - | | + | <td class='row'>ND</td> |
| - | <div style='text-align:justify'> | + | </tr> |
| - | | + | <tr> |
| - | <table align='center' width='100%'> | + | <td class='row'>B0032</td> |
| - | <tr> | + | <td class='row'>3.16</td> |
| - | <td> | + | <td class='row'>~0</td> |
| - | <div style='text-align:center'><div class="thumbinner" style="width:100%;"><img alt="" src="https://static.igem.org/mediawiki/2011/8/88/Caratterizzazione_aiia.JPG" class="thumbimage" width="32%"></a></div></div> | + | <td class='row'>6.7</td> |
| - | </td> | + | <td class='row'>[4.45;10.05]</td> |
| - | </tr> | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0034</td> |
| | + | <td class='row'>2.73</td> |
| | + | <td class='row'>0.23</td> |
| | + | <td class='row'>8.96</td> |
| | + | <td class='row'>[8.27;9.71]</td> |
| | + | </tr> |
| | </table> | | </table> |
| - | </div> | + | <div align="right"><small><a href="#indice" title="">^top</a></small></div> |
| | | | |
| - | <div style='text-align:justify'> | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------AiiA and LuxI-----------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | | | |
| - | <table align='center' width='100%'> | + | <a name='enzymes'></a> |
| - | <tr> | + | <h2>Characterization of enzymes AiiA and LuxI</h2> |
| - | <td> | + | <p align='justify'> LuxI has been characterized through the Biosensor BBa_T9002 (see <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#introduction_to_T9002'>modelling section</a>) using the <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#pTetLuxI'>p<sub>Tet</sub>-RBSx-LuxI-TT</a> measurement systems. <br> |
| - | <div style='text-align:center'><div class="thumbinner" style="width:100%;"><img alt="" src="https://static.igem.org/mediawiki/2011/4/48/Caratterizzazione_luxIN.jpg" class="thumbimage" width="28%"></a></div></div> | + | The parameters V<sub>max</sub>, k<sub>M,LuxI</sub> and α<sub>RBSx</sub> were estimated with a simultaneous fitting of the data collected as described in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#LuxI'>measurement section</a> for the four measurement parts <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#pTetLuxI'>p<sub>Tet</sub>-RBSx-LuxI-TT</a> assayed by <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#T9002'>BBa_T9002 biosensor</a> section. |
| - | </td>
| + | <div align="justify"> |
| - | </tr>
| + | <div class="thumbinner" style="width: 850px;"> <a href="https://static.igem.org/mediawiki/2011/4/40/Luxi_prod.jpg"> <img alt="" src="https://static.igem.org/mediawiki/2011/4/40/Luxi_prod.jpg" class="thumbimage" width="85%" height="50%"></a></div> |
| - | </table>
| + | |
| | </div> | | </div> |
| - | | + | <div align="justify"> |
| - | <div style='text-align:justify'><p>Based on our <a href="#Hypothesis"><span class="toctext"><b><em>HP<sub>3</sub></em></b></span></a> and <a href="#Hypothesis"><span class="toctext"><b><em>HP<sub>4</sub></em></b></span></a>, we are able to determine AiiA and LuxI concentrations, provided we have yet characterized pTet-RBSx contructs<a name='t9002'></a>. In particular, referring to <a href="#Hypothesis"><span class="toctext"><b><em>HP<sub>4</sub></em></b></span></a>, in exponential growth the equilibrium of the enzymes is conserved. Due to a known induction of aTc, the steady-state level per cell can be calculated:</p></div> | + | <p>The estimated parameters for the enzymatic activity of LuxI are reported in the table below:</p> |
| - | | + | |
| - | <div style='text-align:justify'><div class="thumbinner" style="width: 500px;"><img alt="" src="https://static.igem.org/mediawiki/2011/7/74/Aiia_cost.jpg" class="thumbimage" width="120%"></a></div></div>
| + | |
| - | | + | |
| - | <p>Then, as a second step, we monitor in separate experiments HSL synthesis and degradation caused by the activities of the enzymes. In other words, our idea is to control the degradation of HSL versus time. ATc activates pTet and, later, a certain concentration of HSL is introduced. Then, at fixed times, O.D.<sub>600</sub> and HSL concentration are monitored using Tecan and T9002 biosensor.</p><p>For example for AiiA dependent HSL degradation, we have:</p> | + | |
| - | | + | |
| - | <table align='center' width='100%'>
| + | |
| - | <div style='text-align:center'><div class="thumbinner" style="width: 70%;"><img src="https://static.igem.org/mediawiki/2011/9/99/Degradation.jpg" class="thumbimage" width="140%"></a></div></div>
| + | |
| - | </table>
| + | |
| - | | + | |
| - | <br><p>Therefore, considering for a determined promoter-RBSx couple, several induction of aTc and, for each of them, several samples of HSL concentration during time, parameters V<sub>max</sub>, k<sub>M,LuxI</sub>, k<sub>cat</sub> and k<sub>M,AiiA</sub> can be estimated, through numerous iterations of an algorithm implemented in MATLAB.</p>
| + | |
| - | <div align="right"><small><a href="#indice">^top</a></small></div>
| + | |
| - | <br>
| + | |
| - | <br>
| + | |
| - | | + | |
| - | <a name="N"></a><h4> <span class="mw-headline"> <b>N</b> </span></h4>
| + | |
| - | <div style='text-align:justify'>The parameters N<sub>max</sub> and μ can be calculated from the analysis of the OD<sub>600</sub> produced by our MGZ1 culture. In particular, μ is derived as the slope of the log(O.D.<sub>600</sub>) growth curve. Counting the number of cells of a saturated culture would be considerably complicated, so N<sub>max</sub> is determined with a proper procedure. The aim here is to derive the linear proportional coefficient Θ between O.D'.<sub>600</sub> and N: this constant can be estimated as the ratio between absorbance (read from TECAN) and the respective number of CFU on a petri plate. Finally, N<sub>max</sub> is calcultated as Θ*O.D'.<sub>600</sub>
| + | |
| - | <a href="#Pasotti">(<i><b>Pasotti L</b> et al. 2010</i>)</a>.
| + | |
| - | <div align="right"><small><a href="#indice">^top</a></small></div>
| + | |
| | </div> | | </div> |
| - | | + | <div align=center"> |
| - | <br> | + | <table align="center" class='data'> |
| - | <br> | + | <tr> |
| - | | + | <td class='row'><b>V<sub>max</sub></b></td> |
| - | | + | <td class='row'><b>k<sub>M,LuxI</sub></b></td> |
| - | <a name="Degradation_rates"></a><h4> <span class="mw-headline"> <b>Degradation rates</b> </span></h4> | + | <td class='row'><b>α<sub>B0030</sub></b></td> |
| - | <div style='text-align:justify'>The parameters γ<sub>LuxI</sub> and γ<sub>AiiA</sub> are taken from literature since they contain LVA tag for rapid degradation. Instead, approximating HSL kinetics as a decaying exponential, γ<sub>HSL</sub> can be derived as the slope of the log(concentration), which can be monitored through <a href="http://partsregistry.org/Part:BBa_T9002">BBa_T9002</a>. | + | <td class='row'><b>α<sub>B0031</sub></b></td> |
| | + | <td class='row'><b>α<sub>B0032</sub></b></td> |
| | + | <td class='row'><b>α<sub>B0034</sub></b></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>3.56*10<sup>-9</sup></td> |
| | + | <td class='row'>6.87*10<sup>3</sup></td> |
| | + | <td class='row'> 87 </td> |
| | + | <td class='row'>8.5</td> |
| | + | <td class='row'>ND</td> |
| | + | <td class='row'> 252 </td> |
| | + | </tr> |
| | + | </table> |
| | </div> | | </div> |
| - | <div align="right"><small><a href="#indice">^top</a></small></div>
| |
| | <br> | | <br> |
| - | <br> | + | </p> |
| - | | + | </p> |
| - | | + | <p align='justify'> The provided parameters k<sub>M</sub> and V<sub>max</sub> represent the enzymatic activity of LuxI, described by our model. They must not be confused with the operative parameters of the Michaelis-Menten relation. |
| - | <a name="Simulations"></a><h1><span class="mw-headline"> <b>Simulations</b> </span></h1> | + | These synthetic parameters have a great importance, since they can be used in more complicated models in order to predict the behavior of complex circuits. </p> |
| - | <div style='text-align:justify'> | + | <p align='justify'> The AiiA enzyme activity has been characterized under the regulation of p<sub>tet</sub> promoter, assaying its enzymatic activity. </p> |
| - | <p>On a biological level, the ability to control the concentration of a given molecule reveals itself as fundamental in limiting the metabolic burden of the cell; moreover, in the particular case of HSL signalling molecules, this would give the possibility to regulate quorum sensing-based population behaviours. In this section we present some simulations of another circuit, which could validate the concept of the closed-loop model we have discussed so far.</p> | + | <p align='justify'> The parameters k<sub>cat</sub>, k<sub>M,AiiA</sub> and α<sub>RBSx</sub> would have been estimated with a simultaneous fitting of the data collected as described in <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#AiiA'>measurement section</a> for the four measurement parts <a href='https://2011.igem.org/Team:UNIPV-Pavia/Parts/Characterized#pTetAiiA'>p<sub>Tet</sub>-RBSx-AiiA-TT</a> assayed by <a href='https://2011.igem.org/Team:UNIPV-Pavia/Measurements#T9002'>BBa_T9002 biosensor</a> section. |
| - | <p>In order to see that, we implemented and simulated in Matlab an open loop circuit, similar to <b>CTRL+<em>E</em></b>, except for the constitutive production of AiiA.</p> | + | Unfortunately, their estimation revealed difficult.<br> |
| - | | + | In the first experiments with the measurement system <a href="https://static.igem.org/mediawiki/2011/1/11/Pc_aiia.jpg">p<sub>Tet</sub>-RBSx-AiiA-TT</a> in LOW-COPY at pH=7 no degradation of HSL was observed. The collected data are shown in the figure below. HSL degradation is identical in the measurement system and in the negative control after 21 hours. |
| - | <center> | + | <table width='100%'> |
| - | <table> | + | <tr> |
| - | <tr> | + | <td width='50%'><div style="text-align:justify"> |
| - | <td> | + | <div class="thumbinner" width='100%'> <a href="https://static.igem.org/mediawiki/2011/6/69/Aiia_LC_B0030.jpg"> <img alt="" src="https://static.igem.org/mediawiki/2011/6/69/Aiia_LC_B0030.jpg" class="thumbimage" width="100%"></a></div> |
| - | <div style='text-align:justify'><div class="thumbinner" style="width: 500px;"><img src="https://static.igem.org/mediawiki/2011/b/b6/Sim_closed.jpg" class="thumbimage" width="100%"></a></div></div> | + | </div></td> |
| - | </td> | + | <td width='50%'><div style="text-align:justify"> |
| | + | <div class="thumbinner" width='100%'> <a href="https://static.igem.org/mediawiki/2011/6/63/Aiia_LC_B0031.jpg"> <img alt="" src="https://static.igem.org/mediawiki/2011/6/63/Aiia_LC_B0031.jpg" class="thumbimage" width="100%"></a></div> |
| | + | </div></td> |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| - | <td> | + | <td width='50%'><div style="text-align:justify"> |
| - | <div style='text-align:justify'><div class="thumbinner" style="width: 500px;"><img src="https://static.igem.org/mediawiki/2011/4/4b/Sim_open.jpg" class="thumbimage" width="100%"></a></div></div> | + | <div class="thumbinner" width='100%'> <a href="https://static.igem.org/mediawiki/2011/f/f5/Aiia_LC_B0032.jpg"> <img alt="" src="https://static.igem.org/mediawiki/2011/f/f5/Aiia_LC_B0032.jpg" class="thumbimage" width="100%"></a></div> |
| - | </td> | + | </div></td> |
| | + | <td width='50%'><div style="text-align:justify"> |
| | + | <div class="thumbinner" width='100%'> <a href="https://static.igem.org/mediawiki/2011/8/8d/Aiia_LC_B0034.jpg"> <img alt="" src="https://static.igem.org/mediawiki/2011/8/8d/Aiia_LC_B0034.jpg" class="thumbimage" width="100%"></a></div> |
| | + | </div></td> |
| | </tr> | | </tr> |
| | </table> | | </table> |
| - | </center>
| |
| - | </div>
| |
| - | <div align="right"><small><a href="#indice">^top</a></small></div>
| |
| - | <br>
| |
| - | <br>
| |
| | | | |
| - | <a name="Sensitivity_Analysis"></a><h1> <span class="mw-headline"> <b>Sensitivity Analysis of the steady state of enzyme expression in exponential phase</b> </span></h1>
| |
| | | | |
| - | <p>In this paragraph we investigate the theoretical behaviour of our circuit in the cell culture exponential growth phase. According to this, we first derive, under feasible hypotheses, the steady state condition for the enzymes and HSL concentration in that phase. Then we perform a sensitivity analysis relating the output of our system (HSL) to input (aTc) and system parameters.</p> | + | Experiments on these parts gave us the opportunity to characterize only the activity of the enzyme in <em>E. COLI</em> TOP10 in high copy number plasmid, providing only some information about the order of magnitude of the model parameters, which has been designed to work in <em>E. COLI</em> MGZ1 in low copy number plasmid. |
| | + | <div align="right"><small><a href="#indice" title="">^top</a></small></div> |
| | + | |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------RBS---------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | |
| | + | <a name='rbs'></a> |
| | + | <h2>Characterization of the efficiency of RBSs from the community collection</h2> |
| | + | RBSs were used for the fine tuning of CTRL+E. Different experimental conditions were assayed. <br> |
| | + | <br> |
| | + | Estimated efficiencies in pSB4C5 plasmid with -RBSx-mRFP-TT coding sequence under the control of the specified promoter: <br> |
| | + | <br> |
| | + | <div align="center"> |
| | + | <table class='data' width='70%'> |
| | + | <tr> |
| | + | <td class='row'><b>RBS</b></td> |
| | + | <td class='row'><b>eff<sub>p<sub>Lux</sub></sub></b></td> |
| | + | <td class='row'><b>eff<sub>p<sub>Tet</sub></sub></b></td> |
| | + | <td class='row'><b>eff<sub>J23101</sub></b></td> |
| | + | <td class='row'><b>Declared efficiency</b></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0030</td> |
| | + | <td class='row'>0.40</td> |
| | + | <td class='row'>1.6814</td> |
| | + | <td class='row'>2.45</td> |
| | + | <td class='row'>0,6</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0031</td> |
| | + | <td class='row'>0.01</td> |
| | + | <td class='row'>ND</td> |
| | + | <td class='row'>0.04</td> |
| | + | <td class='row'>0,07</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0032</td> |
| | + | <td class='row'>0.19</td> |
| | + | <td class='row'>0.4193</td> |
| | + | <td class='row'>0.40</td> |
| | + | <td class='row'>0,3</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0034</td> |
| | + | <td class='row'>1</td> |
| | + | <td class='row'>1</td> |
| | + | <td class='row'>1</td> |
| | + | <td class='row'>1</td> |
| | + | </tr> |
| | + | </table> |
| | + | </div> |
| | + | <br> |
| | + | <br> |
| | + | Estimated efficiencies in pSB4C5 plasmid with pTet-RBSx-GeneX-TT, with GeneX=mRFP, AiiA or LuxI: <br> |
| | + | <br> |
| | + | <div align="center"> |
| | + | <table class='data' width='70%'> |
| | + | <tr> |
| | + | <td class='row'><b>RBS</b></td> |
| | + | <td class='row'><b>eff<sub>mRFP</sub></b></td> |
| | + | <td class='row'><b>eff<sub>AiiA</sub></b></td> |
| | + | <td class='row'><b>eff<sub>LuxI</sub></b></td> |
| | + | <td class='row'><b>Declared efficiency</b></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0030</td> |
| | + | <td class='row'>1.72</td> |
| | + | <td class='row'>0.53</td> |
| | + | <td class='row'>0.45</td> |
| | + | <td class='row'>0,6</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0031</td> |
| | + | <td class='row'>0.03</td> |
| | + | <td class='row'>0.83</td> |
| | + | <td class='row'>0.028</td> |
| | + | <td class='row'>0,07</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0032</td> |
| | + | <td class='row'>0.37</td> |
| | + | <td class='row'>0.50</td> |
| | + | <td class='row'>N.D.</td> |
| | + | <td class='row'>0.3</td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>B0034</td> |
| | + | <td class='row'>1</td> |
| | + | <td class='row'>1</td> |
| | + | <td class='row'>1</td> |
| | + | <td class='row'>1</td> |
| | + | </tr> |
| | + | </table> |
| | + | </div> |
| | + | <div align="right"><small><a href="#indice" title="">^top</a></small></div> |
| | + | |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!-----------growth---------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | |
| | + | <a name='growth'></a> |
| | + | <h2>Identification of bacterial growth parameters</h2> |
| | | | |
| - | <a name="Steady state of enzyme expression"></a><h2> <span class="mw-headline"> <b>Steady state of enzyme expression</b> </span></h2> | + | |
| | + | <p align='justify'> The bacterial growth curve has been modelled as a logistic curve and is represented by the following equation: <br> |
| | + | <br> |
| | + | dN/dt=N*μ*(N<sub>max</sub>-N)/N<sub>max</sub><br> |
| | + | N(0)=n<sub>0</sub> <br> |
| | + | <br> |
| | + | where μ represents the growth rate of the cells (<em>E. coli</em> MGZ1 in M9 supplemented medium) and N<sub>max</sub> represents the maximum number of cells in the well. For a detailed description of the parameters, see <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#Table_of_parameters'>modelling section</a>. For details on parameters identification, see <a href='https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#N'>identification section.</a> The growth curves in all the performed experiments are measured in O.D.<sub>600</sub>. Since the <em>N</em> species in the model is expressed in <em>cell number</em>, a conversion factor has been estimated. The conversion factor <b>K<sub>O.D.toC.F.U.</sub></b> has been estimated as follows. |
| | + | <ol> |
| | + | <ul> |
| | + | <li>Two cultures C1 and C2 (MGZ1 cells) were grown in 1ml M9 medium till saturation (ON liquid culture, 37°C, 220 rpm).</li> |
| | + | <li>Next morning, both C1 and C2 were diluted in M9 medium with a final volume of 1ml with the following dilution factors: |
| | + | <ul> |
| | + | <li>1:1</li> |
| | + | <li>1:10</li> |
| | + | <li>1:100</li> |
| | + | <li>1:1000</li> |
| | + | </ul> |
| | + | in fresh M9 medium. These cultures were grown for further 1 hour at 37°C, 220 rpm. </li> |
| | + | <li>After 1 hour, O.D.<sub>600</sub> was measured using TECAN microplate reader (don't forget to measure a M9 sample for blanking!)<br> |
| | + | <em>NB: from now on, cultures must be placed in ice to stop cell growth.</em></li> |
| | + | <li>At the same time, proper dilution of the cultures were plated on LB agar plates.<br> |
| | + | <em>NB: All the dilutions are performed moving 100 μl of culture in previously ice-chilled 900 μl fresch M9. 100 μl of the final dilution are plated (It still represents a 1:10 dilution!)</em></li> |
| | + | <li>Plates were grown overnight and next morning C.F.U. were counted.</li> |
| | + | <li>C.F.U. values were corrected by the dilution factor and a linear regression (N vs O.D.<sub>600</sub>) was performed in order to evaluate the conversion factor <b>K<sub>O.D.toC.F.U.</sub></b>. </li> |
| | + | <li><b>K<sub>O.D.toC.F.U.</sub></b> was used as conversion factor to multiply the O.D.<sub>600</sub> value of saturation in the growth curves (~0,5). </li> |
| | + | </ul> |
| | + | </ol> |
| | + | <p>The results are summarized in the table and in the figure below: </p> |
| | + | <center> |
| | + | <table class='data'> |
| | + | <tr> |
| | + | <td class='row'><b> Culture </b></td> |
| | + | <td class='row'><b> O.D.<sub>600</sub> TECAN </b></td> |
| | + | <td class='row'><b> O.D.<sub>600</sub> Spectrophotometer </b></td> |
| | + | <td class='row'><b> C.F.U. </b></td> |
| | + | <td class='row'><b> Dilution Factor (10^) </b></td> |
| | + | <td class='row'><b> N </b></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'> C1 </td> |
| | + | <td class='row'> 0,367950002 </td> |
| | + | <td class='row'> 0,758004416 </td> |
| | + | <td class='row'> 990 </td> |
| | + | <td class='row'> 5 </td> |
| | + | <td class='row'> 990000000 </td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'> C1 </td> |
| | + | <td class='row'> 0,044649998 </td> |
| | + | <td class='row'> 0,091982322 </td> |
| | + | <td class='row'> 141 </td> |
| | + | <td class='row'> 6 </td> |
| | + | <td class='row'> 1410000000 </td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'> C1 </td> |
| | + | <td class='row'> 0,004799999 </td> |
| | + | <td class='row'> 0,009888356 </td> |
| | + | <td class='row'> 136 </td> |
| | + | <td class='row'> 5 </td> |
| | + | <td class='row'> 136000000 </td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'> C1 </td> |
| | + | <td class='row'> 0,000549998 </td> |
| | + | <td class='row'> 0,001133037 </td> |
| | + | <td class='row'> 20 </td> |
| | + | <td class='row'> 6 </td> |
| | + | <td class='row'> 200000000 </td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'> C2 </td> |
| | + | <td class='row'> 0,54840003 </td> |
| | + | <td class='row'> 1,129744917 </td> |
| | + | <td class='row'> 165 </td> |
| | + | <td class='row'> 4 </td> |
| | + | <td class='row'> 16500000 </td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'> C2 </td> |
| | + | <td class='row'> 0,058200002 </td> |
| | + | <td class='row'> 0,119896339 </td> |
| | + | <td class='row'> 23 </td> |
| | + | <td class='row'> 5 </td> |
| | + | <td class='row'> 23000000 </td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'> C2 </td> |
| | + | <td class='row'> 0,008700002 </td> |
| | + | <td class='row'> 0,017922652 </td> |
| | + | <td class='row'> 251 </td> |
| | + | <td class='row'> 3 </td> |
| | + | <td class='row'> 2510000 </td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'> C2 </td> |
| | + | <td class='row'> 0,000100002 </td> |
| | + | <td class='row'> 0,000206011 </td> |
| | + | <td class='row'> 24 </td> |
| | + | <td class='row'> 4 </td> |
| | + | <td class='row'> 2400000 </td> |
| | + | </tr> |
| | + | </table> |
| | + | </center> |
| | + | <br> |
| | + | </p> |
| | + | <center> |
| | + | <img alt="" src="https://static.igem.org/mediawiki/2011/3/35/UNIPV_ODvsCFU.png" width="90%"> |
| | + | </center> |
| | + | <p>The estimation of μ parameter was performed by determining the slope of the logarithmic curve of O.D.<sub>600</sub> in exponential phase. Exponential phase was determined by visual inspection as the linear phase of the logarithmic curve of O.D.<sub>600</sub>. <br> |
| | + | <br> |
| | + | The estimated parameters are summarized in the table below: <br> |
| | + | </p> |
| | + | <center> |
| | + | <table width='50%' class='data'> |
| | + | <tr> |
| | + | <td class='row'><b>N<sub>max</sub> [cell number]</b></td> |
| | + | <td class='row'><b>μ [min<sup>-1</sup>]</b></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>1*10<sup>9</sup></td> |
| | + | <td class='row'>0.004925</td> |
| | + | </tr> |
| | + | </table> |
| | + | </center> |
| | + | <p> </p> |
| | + | <p>The reported value of μ corresponds to a doubling time of 142 minutes. </p> |
| | + | <div align="right"><small><a href="#indice" title="">^top</a></small></div> |
| | + | |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------HSL---------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | |
| | + | <a name='HSL'></a> |
| | + | <h2>Estimation of the spontaneous degradation of HSL in M9 medium and in cultures at different pH values</h2> |
| | + | <p align='justify'> In order to estimate the spontaneous degradation rate of HSL in M9 medium and in a culture of MGZ1 cells as a function of pH, two simple tests have been performed.<br> |
| | + | Two different M9 media were prepared, one with the nominal pH (7.0) and one with pH=6.0.<br> |
| | + | These media, now named respectively M9<sub>pH 7</sub> and M9<sub>pH 6</sub>, were added with a known concentration of HSL (100 nM) and then incubated at 37°C, 220 rpm (NB: the media were not infected with any culture but the standard growth conditions were reproduced). The amount of HSL present in the medium was assayed through the BBa_T9002 biosensor at 4 time points: <br> |
| | + | <ul> |
| | + | <li>t=0 h;</li> |
| | + | <li>t=1 h;</li> |
| | + | <li>t=2 h;</li> |
| | + | <li>t=4 h;</li> |
| | + | <li>t=8 h;</li> |
| | + | </ul> |
| | + | <p>The obtained time series of HSL amounts were processed to evaluate the time constant governing the dynamic of HSL degradation, supposing an exponential decay. The results are reported in the table below: </p> |
| | + | <center> |
| | + | <table class='data'> |
| | + | <tr> |
| | + | <td class='row'></td> |
| | + | <td class='row'><b>t<sub>1/2</sub><sup>*</sup> [h]</b></td> |
| | + | <td class='row'><b>γ<sub>HSL</sub><sup>**</sup> [h<sup>-1</sup>]</b></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'><b>M9<sub>pH 6</sub></b></td> |
| | + | <td class='row'>32 </td> |
| | + | <td class='row'>0.022 </td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'><b>M9<sub>pH 7</sub></b></td> |
| | + | <td class='row'>8 </td> |
| | + | <td class='row'>0.087 </td> |
| | + | </tr> |
| | + | </table> |
| | + | </center> |
| | + | <p> The described experiment was repeated with a MGZ1 culture in order to evaluate the effect of culture on HSL stability. The estimated values are reported in the table below:</p> |
| | + | <center> |
| | + | <table class='data'> |
| | + | <tr> |
| | + | <td class='row'></td> |
| | + | <td class='row'><b>t<sub>1/2</sub><sup>*</sup> [h]</b></td> |
| | + | <td class='row'><b>γ<sub>HSL</sub><sup>**</sup> [h<sup>-1</sup>]</b></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'><b>Culture<sub>pH 6</sub></b></td> |
| | + | <td class='row'>∞ </td> |
| | + | <td class='row'>0 </td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'><b>Culture<sub>pH 7</sub></b></td> |
| | + | <td class='row'>19 </td> |
| | + | <td class='row'>0.037 </td> |
| | + | </tr> |
| | + | </table> |
| | + | </center> |
| | + | <p>It is evident from the reported data that the spontaneous degradation of HSL is negligible when CTRL+E is implemented in MGZ1 cells grown in M9 medium with pH=6.0 and pH=7.0.<br> |
| | + | Thus in the simulations we set γ<sub>HSL</sub>=0; </p> |
| | + | </p> |
| | + | <div align="right"><small><a href="#indice" title="">^top</a></small></div> |
| | + | |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!-------------t9002--------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | <!--------------------------------> |
| | + | |
| | + | <a name='t9002'></a> |
| | + | <h2>Characterization of BBa_T9002 biosensor</h2> |
| | + | As described in the <a href="https://2011.igem.org/Team:UNIPV-Pavia/Project/Modelling#introduction_to_T9002">modelling section</a>, BioBrick <a href="http://partsregistry.org/Part:BBa_T9002">BBa_T9002</a> is an HSL biosensor, which provides a non linear relationship between HSL input and S<sub>cell</sub> output. More precisely, the characteristic sigmoidal curve requires synthetic parameters for its accurate identification. These are the minimum and maximum values, the swtich point (i.e., the curve inflection point), and the upper and lower boundaries of linearity. This biosensor revealed greatly reliable, providing measurement repeatability and minimal experimental noise. Referring to its activation formula, the calibration curve is shown below.<br> |
| | + | <br> |
| | + | <div style='text-align:center'> |
| | + | <div class="thumbinner" style="width:100%;"> <img alt="" src="https://static.igem.org/mediawiki/2011/5/50/Activation_T9002.jpg" class="thumbimage" width="47%" height="50%"></a></div> |
| | + | </div> |
| | + | <br> |
| | + | <div style='text-align:center'> |
| | + | <div class="thumbinner" style="width:100%;"> <img alt="" src="https://static.igem.org/mediawiki/2011/4/4e/T9002_activation.jpg" class="thumbimage" width="100%"></a></div> |
| | + | </div> |
| | + | <center> |
| | + | <table width='50%' class='data'> |
| | + | <tr> |
| | + | <td class='row'><b>Minimum [S<sub>cell</sub>]</b></td> |
| | + | <td class='row'><b>Maximum [S<sub>cell</sub>]</b></td> |
| | + | <td class='row'><b>Switch point [nM]</b></td> |
| | + | <td class='row'><b>Lower boundary of linearity [nM]</b></td> |
| | + | <td class='row'><b>Upper boundary of linearity [nM]</b></td> |
| | + | </tr> |
| | + | <tr> |
| | + | <td class='row'>17.31</td> |
| | + | <td class='row'>739.4</td> |
| | + | <td class='row'>1.39</td> |
| | + | <td class='row'>0.38</td> |
| | + | <td class='row'>5.07</td> |
| | + | </tr> |
| | + | </table> |
| | + | </center> |
| | + | <br> |
| | + | <br> |
| | + | In order to determine the threshold sensitivity of T9002 biosensor, experiments were performed with several HSL inductions minimally interspaced in the region of low detectability. Hypothesizing that the inducer is 1:20 diluted (as for all of our tests), the minimum detectable HSL concentration is 3 nM. |
| | + | <div align="right"><small><a href="#indice" title="">^top</a></small></div> |
| | <br> | | <br> |
| - |
| |
| - | <p> Based on our <a href="#Hypothesis"><span class="toctext"><b><em>HP<sub>4</sub></em></b></span></a>, we can formulate the steady state expressions during the exponential growth phase. Adding other considerations about the involved processes, it is possible to add further simplifications to the steady state equations. In particular, one concern relates to the number of cells N (in the order of 3*10^3), which is far lower than N<sub>max</sub> (10^9). The other pertains to γ*HSL parameter, whch can be neglected compared to the other two terms of the third equation. Based on this assumptions, equation (4) of the system becomes dN/dt=μN. Moreover, from equation (3), after having removed the third term, we can simplify the N parameter, since it is common to the remaining two terms. On a biological point of view, this implies that AiiA, LuxI and HSL undergo only minor changes through time, thereby allowing to derive their steady state expressions:</p>
| |
| - |
| |
| - | <table align='center' width='100%'>
| |
| - | <tr>
| |
| - | <td>
| |
| - | <div style='text-align:center'><div class="thumbinner" style="width: 100%;"><a href="" class="image"><img alt="File:LuxI SS.jpg" src="https://static.igem.org/mediawiki/2011/3/32/LuxI_SS.jpg" class="thumbimage" width="83%"></a></div></div>
| |
| - | </td>
| |
| - | </tr>
| |
| - | </table>
| |
| - |
| |
| - | <p> The first equation is independent from the second and third ones, enabling us to directly determine LuxI steady state expression during the exponential growth phase. On the contrary, second and third equations depend each other in defining the value of AiiA and HSL respectively, because the former is a function of HSL, while the latter is a function of AiiA. So we could resolve a system of two equations, first by expliciting one of the two variables with respect to the other, and then substituting its expression in order to determine the other variable possible values. This would bring a complex mathematical formulation, which is not helpful in understanding the influence of the various model parameters on the output HSL.
| |
| - | On the other hand, AiiA and HSL values can also be graphically determined from the intersection of the curves derived from these two equations, if we explicit HSL as a function of AiiA (or, alternatively, AiiA as a function of HSL). It is easy to discover that these two curves represent rectangular hyperbolae (the first one only under a simple approximation, explained below) whose tails intersect each other at a particular point, corresponding to the searched values for AiiA and HSL.</p>
| |
| - | <div>For a rectangular hyperbola (RH), we have:</div>
| |
| - |
| |
| - | <table align='center' width='100%'>
| |
| - | <tr>
| |
| - | <td>
| |
| - | <div style='text-align:center'><div class="thumbinner" style="width: 100%;"><a href="" class="image"><img alt="File:UNIPV Rectangular hyperbola general.jpg" src="https://static.igem.org/mediawiki/2011/7/75/UNIPV_Rectangular_hyperbola_general.jpg" class="thumbimage" width="14%"></a></div></div>
| |
| - | </td>
| |
| - | </tr>
| |
| - | </table>
| |
| - |
| |
| - | <p>centered at O(-d/c;a/c), with the vertical asymptote x=-d/c and the horizontal asymptote y=a/c</p>
| |
| - | <p>From equation 2, we have:</p>
| |
| - |
| |
| - | <table align='center' width='100%'>
| |
| - | <tr>
| |
| - | <td>
| |
| - | <div style='text-align:center'><div class="thumbinner" style="width: 100%;"><a href="" class="image"><img alt="File:UNIPV eta root HSL.jpg" src="https://static.igem.org/mediawiki/2011/3/3a/UNIPV_eta_root_HSL.jpg" class="thumbimage" width="62%"></a></div></div>
| |
| - | </td>
| |
| - | </tr>
| |
| - | </table>
| |
| - |
| |
| - | <p> We can introduce the simplification to remove the ηplux exponent to the entire expression in the right hand side of the equation, thereby obtaining a rectangular hyperbola; even if this leads to a slight change in the curve behaviour, it allows to more clearly understand the relation between HSL and AiiA. As pertains to equation 3, its steady state approximation during the exponential growth is more immediately identifiable as a rectangular hyperbola. Below the two RHs equations are provided, togheter with the table of parameters.</p>
| |
| - |
| |
| - | <table align='center' width='100%'>
| |
| - | <tr>
| |
| - | <td>
| |
| - | <div style='text-align:center'><div class="thumbinner" style="width: 100%;"><a href="" class="image"><img alt="File:RH1 UNIPV HSL.jpg" src="https://static.igem.org/mediawiki/2011/1/11/RH1_UNIPV_HSL.jpg" class="thumbimage" width="50%"></a></div></div>
| |
| - | </td>
| |
| - | </tr>
| |
| - | </table>
| |
| - |
| |
| - |
| |
| - | <table align='center' width='100%'>
| |
| - | <tr>
| |
| - | <td>
| |
| - | <div style='text-align:center'><div class="thumbinner" style="width: 100%;"><a href="" class="image"><img alt="File:RH2 UNIPV HSL.jpg" src="https://static.igem.org/mediawiki/2011/4/4b/RH2_UNIPV_HSL.jpg" class="thumbimage" width="70%"></a></div></div>
| |
| - | </td>
| |
| - | </tr>
| |
| - | </table>
| |
| | <br> | | <br> |
| - |
| |
| - |
| |
| - | <center>
| |
| - | <table class="data">
| |
| - | <tr>
| |
| - | <td class="row"><b>Parameter</b></td>
| |
| - | <td class="row"><b>RH1</b></td>
| |
| - | <td class="row"><b>RH2</b></td>
| |
| - | </tr>
| |
| - |
| |
| - | <tr>
| |
| - | <td class="row">a</sub></sub></td>
| |
| - | <td class="row">(γ<sub>AiiA</sub>+μ)*(k<sub>pLux</sub>)<sup>ηpLux</td>
| |
| - | <td class="row"><span style="text-decoration:overline;" > V</span><sub>LuxI</sub></td>
| |
| - | </tr>
| |
| - |
| |
| - | <tr>
| |
| - | <td class="row">b</td>
| |
| - | <td class="row">-α<sub>pLux</sub>*δ<sub>pLux</sub>*(k<sub>pLux</sub>)<sup>ηpLux</td>
| |
| - | <td class="row"><span style="text-decoration:overline;" > V</span><sub>LuxI</sub>*k<sub>M,LuxI</sub></td>
| |
| - | </tr>
| |
| - |
| |
| - | <tr>
| |
| - | <td class="row">c</sub></sub></td>
| |
| - | <td class="row">-γ<sub>AiiA</sub>+μ</td>
| |
| - | <td class="row">k<sub>cat</sub>*<span style="text-decoration:overline;" >AiiA</span></td>
| |
| - | </tr>
| |
| - |
| |
| - | <tr>
| |
| - | <td class="row">d</td>
| |
| - | <td class="row">α<sub>pLux</sub></td>
| |
| - | <td class="row">0</td>
| |
| - | </tr>
| |
| - |
| |
| - | <tr>
| |
| - | <td class="row">Horizontal asymptote</td>
| |
| - | <td class="row">a/c=(k<sub>pLux</sub>)<sup>ηpLux</sup></td>
| |
| - | <td class="row"><span style="text-decoration:overline;" > V</span><sub>LuxI</sub>/k<sub>cat</sub></td>
| |
| - |
| |
| - | </tr>
| |
| - |
| |
| - | <tr>
| |
| - | <td class="row">Vertical asymptote</td>
| |
| - | <td class="row">-d/c=α<sub>pLux</sub>/(γ<sub>AiiA</sub>+μ)</td>
| |
| - | <td class="row">0</td>
| |
| - | </tr>
| |
| - | </table>
| |
| - |
| |
| - | </center>
| |
| - |
| |
| - |
| |
| - | <a name="Sensitivity analysis"></a><h2>
| |
| - | <span class="mw-headline"> <b>Sensitivity analysis</b> </span></h2>
| |
| - |
| |
| - | <p> Now, it is interesting to conduct some qualitative and quantitative considerations about our system sensitivity to its parameters and aTc input signal.</p>
| |
| - | <div> First of all, we analyze how the HSL output can be regulated by changing the characteristics of our RHs.</div><div>Referring to the first rectangular hyperbola, we recognize that its vertical asymptote could be varied by changing α<sub>p<sub>Lux</sub></sub></sub></sub> value (assuming fixed γ<sub>AiiA</sub> and μ). In particular, thanks to the four Plux-RBSx constructs realized, we can vary α<sub>p<sub>Lux</sub></sub></sub></sub> more than a hundred factor. This can significantly shift the vertical asymptote, bringing this first RH farther or nearer the second one (whose vertical asymptote is the ordinate axis), thereby providing an intersection at higher AiiA and lower HSL values, or vice versa. The following two figures highlight this aspect.</div>
| |
| - |
| |
| - | <table align='center' width='100%'>
| |
| - | <div style='text-align:center'><div class="thumbinner" style="width: 100%;"><a href="File:Iperbole eq 3 as ver2.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/4/45/Iperbole_eq_3_as_ver2.jpg" class="thumbimage" width="100%"></a></div></div>
| |
| - | </table>
| |
| - |
| |
| - | <table align='center' width='100%'>
| |
| - | <div style='text-align:center'><div class="thumbinner" style="width: 100%;">
| |
| - | <a href="File:UNIPV inters RH ver alfa plux acceptable values2.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/9/97/UNIPV_inters_RH_ver_alfa_plux_acceptable_values2.jpg" class="thumbimage" width="100%"></a></div></div>
| |
| - | </table>
| |
| - |
| |
| - |
| |
| - | <p>From the above figures, it is also clear that HSL steady state value is not very sensitive to α<sub>p<sub>Lux</sub></sub></sub></sub>, at least when this parameter presents values greater than unity, because this brings the two curves to intersect in their low slope regions.</p>
| |
| - | <p>Referring to the second RH, the only adjustable asymptote is the horizontal one, that we can move upward or downward by altering VLuxI, which indirectly depends on aTc.</p>
| |
| - |
| |
| - | <table align='center' width='100%'>
| |
| - | <div style='text-align:center'><div class="thumbinner" style="width: 100%;"><a href="File:UNIPV RH2 eq2 as or corrected2.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/0/04/UNIPV_RH2_eq2_as_or_corrected2.jpg" class="thumbimage" width="95%"></a></div></div>
| |
| - | </table>
| |
| - |
| |
| - | <p>A further deepening in the exponential phase analysis involves determining the characteristics of the input-output relation. Our closed loop system can be realized with two alternative purposes in mind:</p>
| |
| - | <ol>
| |
| - | <li> realizing a circuit able to adapt HSL output depending on aTc input concentration. This requires a good sensitivity between input and output.</li>
| |
| - | <li> designing a robust HSL concentration controller, which is immune to the input noise and offers a constant and defined amount of HSL. In this case HSL level should be appropriately tuned during the design stage, by choosing the correct strength of promoter-RBSx complexes.</li>
| |
| - | </ol>
| |
| - |
| |
| - | <p>Now,again considering the system of equations, it is easy to observe that HSL dependence on aTc input passes through two Hill equations. The former describes aTc driven LuxI synthesis, while the latter models LuxI dependent HSL synthesis rate. Therefore, in order to achieve a high aTc sensitivity, it is advisable to tune aTc and LuxI levels so that they place outside the saturation regions of their Hill curves. In this regard, it is possible to determine a closed form expression relating HSL to aTc, if we hypothesize that both aTc and LuxI are far lower than their respective half-saturation constant (k_ptet and Km), thus simplifying the Hills with a first order relation:</p>
| |
| - |
| |
| | <br> | | <br> |
| | | | |
| - | <table align='center' width='100%'> | + | </html> |
| - | <div style='text-align:center'><div class="thumbinner" style="width: 100%;"><a href="File:UNIPV Sensitivity atc2.jpg" class="image"><img alt="" src="https://static.igem.org/mediawiki/2011/3/34/UNIPV_Sensitivity_atc2.jpg" class="thumbimage" width="75%"></a></div></div>
| + | |
| - | </table>
| + | |
| | | | |
| | | | |
| - | <a name="References"></a><h1><span class="mw-headline"> <b>References</b> </span></h1>
| |
| - | <div style='text-align:justify'>
| |
| - |
| |
| - | <ol type='1'>
| |
| - |
| |
| - | <a name="Braun"><li></a>Braun D, Basu S, Weiss R (2005) <b>Parameter estimation for two synthetic gene networks: a case study </b>
| |
| - | <i>ICASSP '05 </i> 5:v/769-v/772. </li> <br>
| |
| - |
| |
| - | <a name="Canton"><li></a>Canton B, Labno A, Endy D. (2008) <b>Refinement and standardization of synthetic biological parts and devices. </b> <i> Nat Biotechnol. </i> 26(7):787-93. </li> <br>
| |
| - |
| |
| - | <a name="Danino"><li></a>Danino T, Mondragón-Palomino O, Tsimring L et al. (2010) <b>A synchronized quorum of genetic clocks. </b> <i>Nature. </i> 463(7279):326-30. </li> <br>
| |
| - |
| |
| - | <a name="Endler"><li></a>Endler L, Rodriguez N, Juty N et al. (2009) <b>Designing and encoding models for synthetic biology. </b> <i>J. R. Soc. Interface </i> 6:S405-S417. </li> <br>
| |
| - |
| |
| - | <a name="Kelly"><li></a>Kelly JR, Rubin AJ, Davis JH et al. (2009) <b>Measuring the activity of BioBrick promoters using an in vivo reference standard.</b> <i> J. Biol. Eng.</i> 3:4.</li> <br>
| |
| - |
| |
| - | <a name="Pasotti"><li></a>Pasotti L, Quattrocelli M, Galli D et al. (2011) <b>Multiplexing and demultiplexing logic functions for computing signal processing tasks in synthetic biology. </b>
| |
| - | <i>Biotechnol. J. </i>6(7):784-95. </li> <br>
| |
| - |
| |
| - | <a name="Salis"><li></a> Salis H M, Mirsky E A, Voight C A (2009)<b> Automated design of synthetic ribosome binding sites to control protein expression. </b> <i>Nat. Biotechnol.</i>27:946-950.
| |
| - | </li><br>
| |
| - |
| |
| - |
| |
| - | </ol>
| |
| - |
| |
| - | </div>
| |
| - | </div>
| |
| - |
| |
| - | </html>
| |
| | | | |
| | {{end}} | | {{end}} |
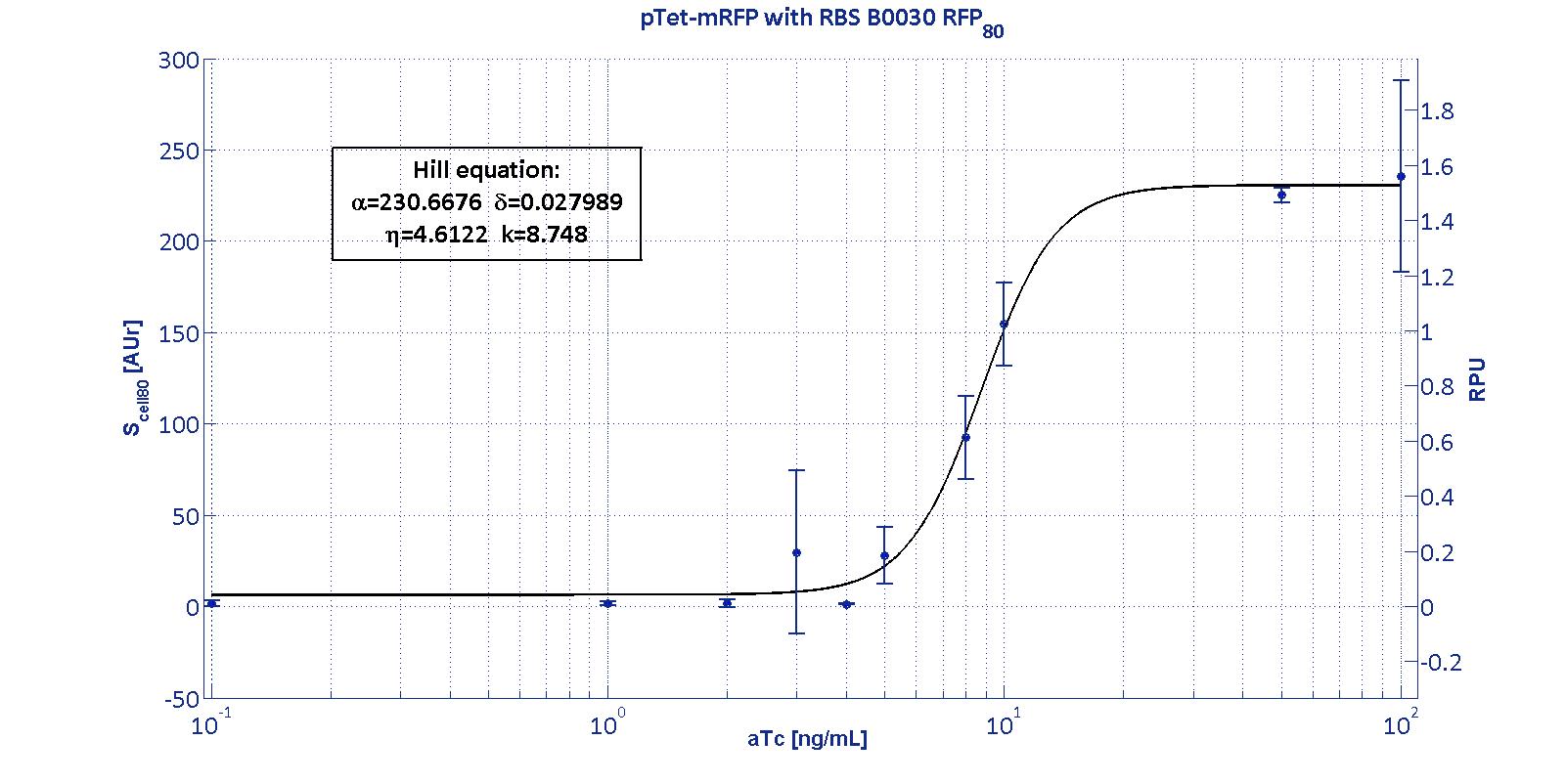

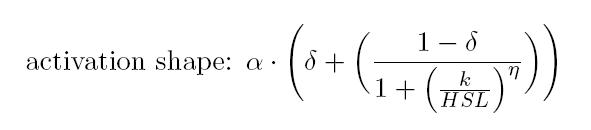
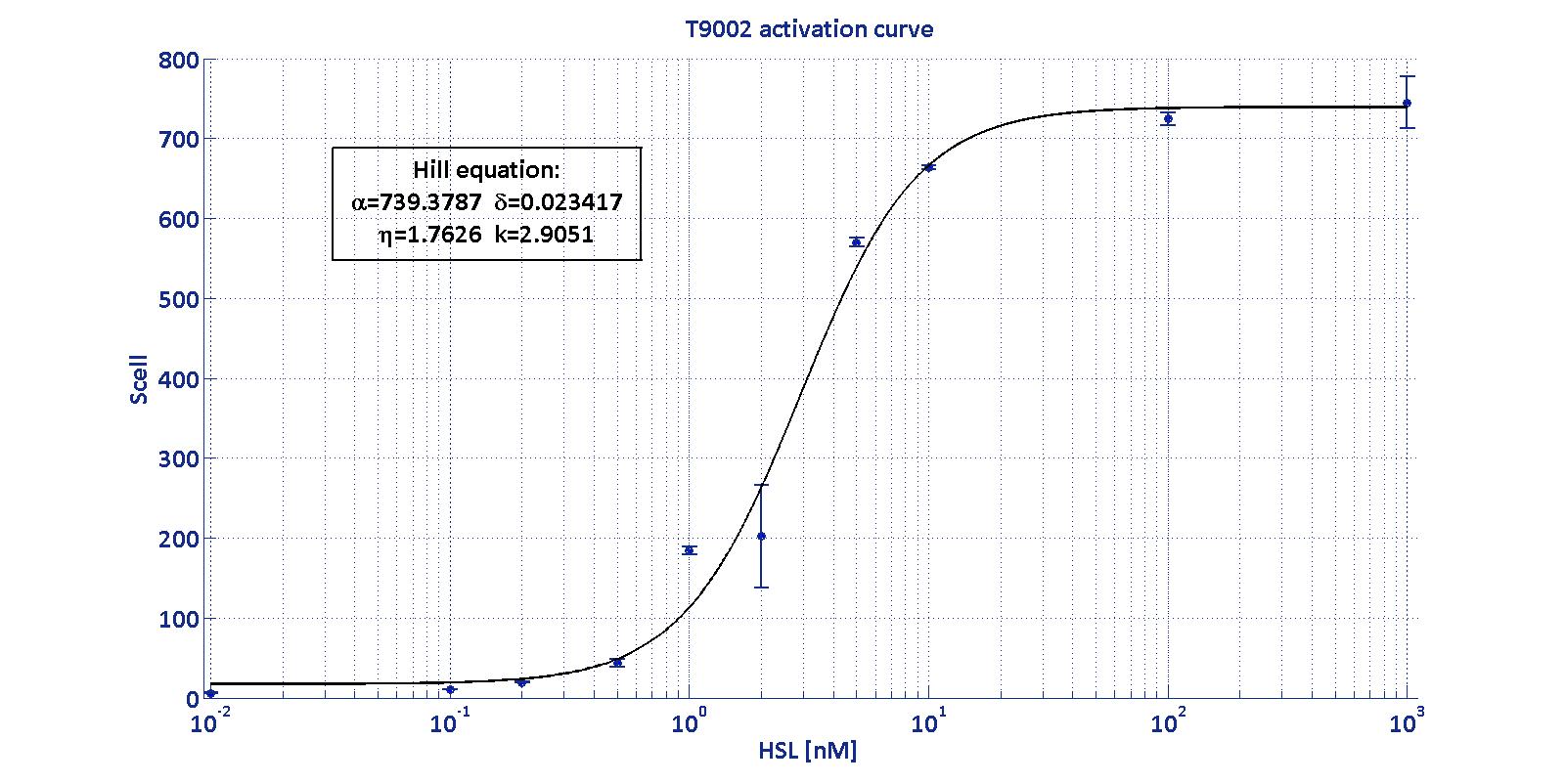
 "
"










