Team:Bielefeld-Germany/Results/S-Layer/Guide/3a
From 2011.igem.org
| (11 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Bielefeld_2011_Header}} | {{Bielefeld_2011_Header}} | ||
| + | <html><img src="https://static.igem.org/mediawiki/2011/2/28/Bielefeld-header-guide.png"/><p></p></html> | ||
| + | =<html><div style="line-height:1.2em;">Easy and small scale expression of S-layer protein under control of T7 promoter</div></html>= | ||
| - | + | [[Image:IGEM-Bielefeld2011-Shaker.JPG|400px|thumb|right|Shaking flasks on a Kuhner Lab-Shaker LS-X]] | |
| - | [ | + | The expression of S-layer proteins is stressful for ''Escherichia coli''. So using ''E. coli'' [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ KRX] to express genes under the control of a T7 promoter is an easy way to overexpress your proteins and seperate growth and production phase. This strain carries a T7 polymerase gene under the control of a rhamnose promoter in its genome and is also optimized for cloning so you do not have to transform your plasmids after assembly in e.g. TOP10, isolate them and bring them in an expression strain like BL21(DE3) - you can go with a single transformation step from assembly to production. The rhamnose promoter is a tightly controlled promoter (compared to arabinose or lactose promoter) and is inhibited by glucose. Using glucose and L-rhamnose supplemented LB medium leads to an autoinduction of the rhamnose promoter when glucose is completely metabolized by the cells. L-rhamnose can not be metabolized by the cells but has the same effect like D-rhamnose on the rhamnose promoter. |
| - | + | Summarized: Use glucose, L-rhamnose and antibiotic supplemented LB medium, put it in a shaking flask, add your ''E. coli'' KRX cells carrying your S-layer fusion protein under the control of a T7 promoter, put the shaking flask on a shaker at 37 °C and then just wait! The cells will grow until the glucose is depleted (OD<sub>600</sub> ~ 1 when inoculating with OD<sub>600</sub> ~ 0.1) and the expression of the S-layer will start. Stop the cultivation after about 8 - 10 h and harvest your cells by centrifugation. | |
| - | + | ||
| - | Summarized: Use glucose, L-rhamnose and antibiotic supplemented LB medium, put it in a shaking flask, add your ''E. coli'' KRX cells carrying your S-layer fusion protein under the control of a T7 promoter, put the shaking flask on a shaker at 37 °C and then just wait! The cells will grow until the glucose is | + | |
Want to know how to continue? Then read [[Team:Bielefeld-Germany/Results/S-Layer/Guide/4a | how to disrupt your cells here]]. | Want to know how to continue? Then read [[Team:Bielefeld-Germany/Results/S-Layer/Guide/4a | how to disrupt your cells here]]. | ||
| - | To show that this really works | + | To show that this really works that easy: Look at the following figures displaying the expression of the fluorescent S-layer fusion protein SgsE | mCitrine using the autoinduction protocol. |
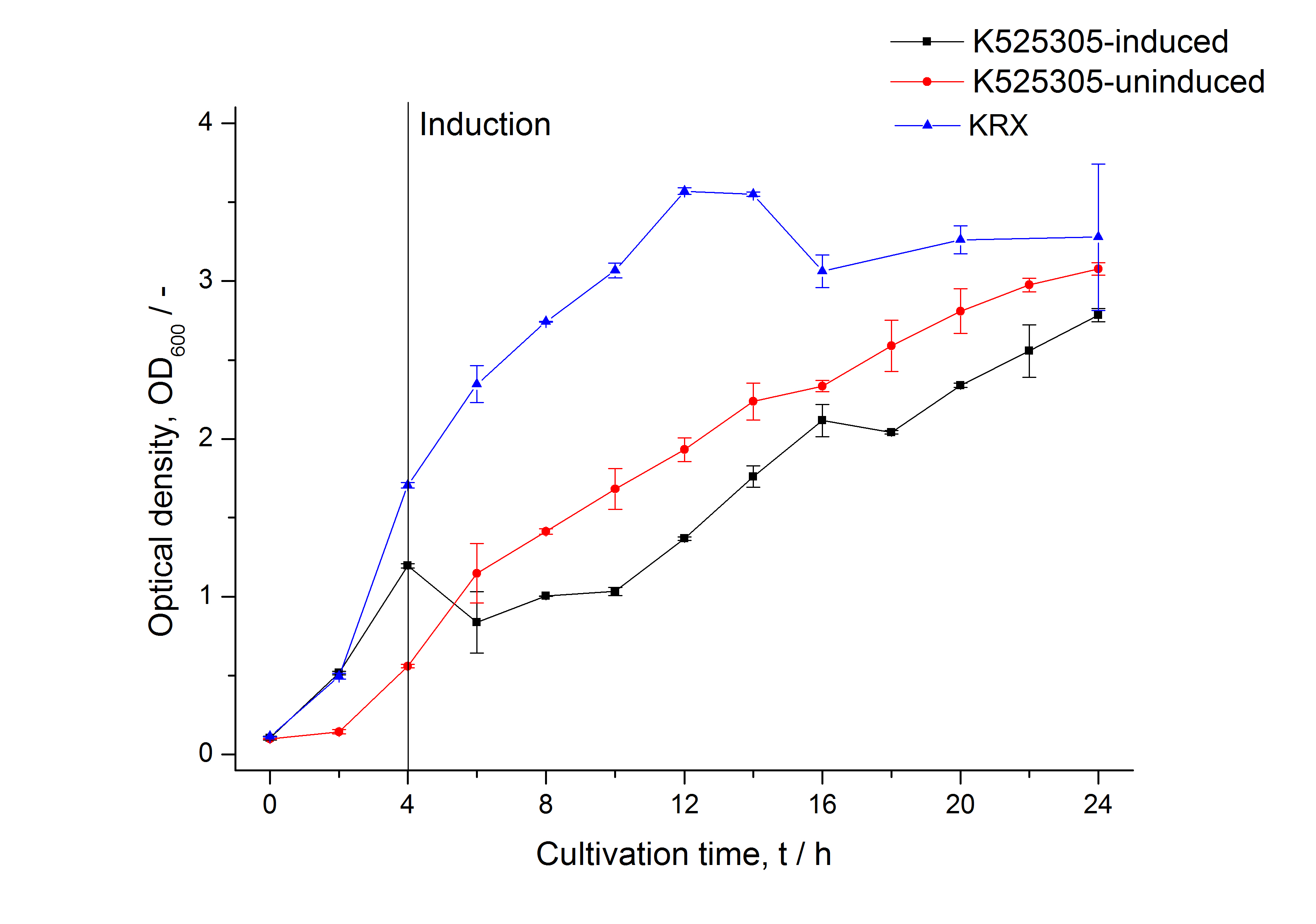
[[Image:Bielefeld_2011_305_Growthcurve.png|430px|left|thumb| '''Figure 1: Growth curve of ''E. coli'' KRX expressing the fusion protein of SgsE and mCitrine with and without induction, cultivated at 37 °C in autoinduction medium with and without inductor, respectively. A curve depicting KRX wildtype is shown for comparison. After induction at approximately 4 h the OD<sub>600</sub> of the induced <partinfo>K525305</partinfo> visibly drops when compared to the uninduced culture. Both cultures grow significantly slower than KRX wildtype probably due to a leaky promoter and metabolic stress by the high copy plasmid.''']] | [[Image:Bielefeld_2011_305_Growthcurve.png|430px|left|thumb| '''Figure 1: Growth curve of ''E. coli'' KRX expressing the fusion protein of SgsE and mCitrine with and without induction, cultivated at 37 °C in autoinduction medium with and without inductor, respectively. A curve depicting KRX wildtype is shown for comparison. After induction at approximately 4 h the OD<sub>600</sub> of the induced <partinfo>K525305</partinfo> visibly drops when compared to the uninduced culture. Both cultures grow significantly slower than KRX wildtype probably due to a leaky promoter and metabolic stress by the high copy plasmid.''']] | ||
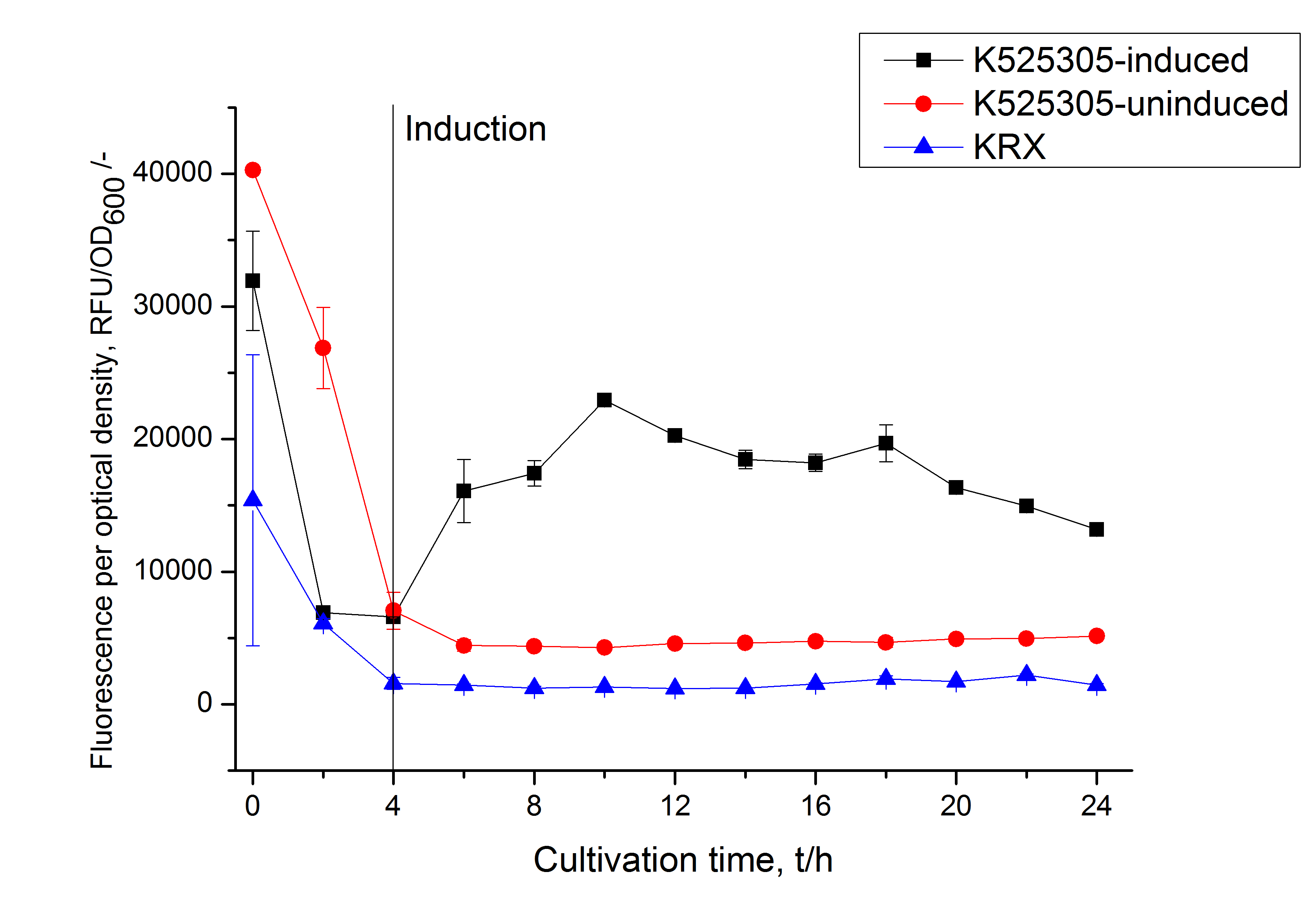
[[Image:Bielefeld_2011_305_RFU_OD.png|430px|left|thumb| '''Figure 2: RFU to OD<sub>600</sub> ratio of ''E. coli'' KRX expressing the fusion protein of SgsE and mCitrine with and without induction. A curve depicting KRX wildtype is shown for comparison. After induction at approximately 4 h the RFU to OD<sub>600</sub> ratio starts to rise in the induced culture. Compared to the uninduced culture the ratio is roughly four times higher. The KRX wildtype shows no variation in the RFU to OD<sub>600</sub> ratio.''']] | [[Image:Bielefeld_2011_305_RFU_OD.png|430px|left|thumb| '''Figure 2: RFU to OD<sub>600</sub> ratio of ''E. coli'' KRX expressing the fusion protein of SgsE and mCitrine with and without induction. A curve depicting KRX wildtype is shown for comparison. After induction at approximately 4 h the RFU to OD<sub>600</sub> ratio starts to rise in the induced culture. Compared to the uninduced culture the ratio is roughly four times higher. The KRX wildtype shows no variation in the RFU to OD<sub>600</sub> ratio.''']] | ||
| + | |||
| + | <html> | ||
| + | <div style="text-align:center; width:535px; margin-left:auto; margin-right:auto;"> | ||
| + | <img id="Image-Maps_8201110281849269" src="https://static.igem.org/mediawiki/2011/0/0c/Bielefeld-Germany-2011-Navigationbar_a.png" usemap="#Image-Maps_8201110281849269" border="0" width="535" height="82" alt="" /> | ||
| + | <map id="_Image-Maps_8201110281849269" name="Image-Maps_8201110281849269"> | ||
| + | <area shape="rect" coords="22,12,170,62" href="https://2011.igem.org/Team:Bielefeld-Germany/Results/S-Layer/Guide/2" alt="Previous page" title="Previous page" /> | ||
| + | <area shape="rect" coords="182,7,330,57" href="https://2011.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Results/S-Layer/Guide" alt="Return to beginning" title="Return to beginning" /> | ||
| + | <area shape="rect" coords="353,10,501,60" href="https://2011.igem.org/Team:Bielefeld-Germany/Results/S-Layer/Guide/4a" alt="Next page" title="Next page" /> | ||
| + | </map> | ||
| + | </div> | ||
| + | </html> | ||
Latest revision as of 02:48, 29 October 2011


Easy and small scale expression of S-layer protein under control of T7 promoter
The expression of S-layer proteins is stressful for Escherichia coli. So using E. coli [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/single-step-_krx_-competent-cells/ KRX] to express genes under the control of a T7 promoter is an easy way to overexpress your proteins and seperate growth and production phase. This strain carries a T7 polymerase gene under the control of a rhamnose promoter in its genome and is also optimized for cloning so you do not have to transform your plasmids after assembly in e.g. TOP10, isolate them and bring them in an expression strain like BL21(DE3) - you can go with a single transformation step from assembly to production. The rhamnose promoter is a tightly controlled promoter (compared to arabinose or lactose promoter) and is inhibited by glucose. Using glucose and L-rhamnose supplemented LB medium leads to an autoinduction of the rhamnose promoter when glucose is completely metabolized by the cells. L-rhamnose can not be metabolized by the cells but has the same effect like D-rhamnose on the rhamnose promoter.
Summarized: Use glucose, L-rhamnose and antibiotic supplemented LB medium, put it in a shaking flask, add your E. coli KRX cells carrying your S-layer fusion protein under the control of a T7 promoter, put the shaking flask on a shaker at 37 °C and then just wait! The cells will grow until the glucose is depleted (OD600 ~ 1 when inoculating with OD600 ~ 0.1) and the expression of the S-layer will start. Stop the cultivation after about 8 - 10 h and harvest your cells by centrifugation.
Want to know how to continue? Then read how to disrupt your cells here.
To show that this really works that easy: Look at the following figures displaying the expression of the fluorescent S-layer fusion protein SgsE | mCitrine using the autoinduction protocol.

 "
"

