Team:Bielefeld-Germany/Results/S-Layer/Guide/1
From 2011.igem.org
(→Planning a nanobiotechnological device) |
|||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Bielefeld_2011_Header}} | {{Bielefeld_2011_Header}} | ||
| - | + | <html><img src="https://static.igem.org/mediawiki/2011/2/28/Bielefeld-header-guide.png"/><p></p></html> | |
=Planning a nanobiotechnological device= | =Planning a nanobiotechnological device= | ||
| + | [[Image:IGEM_Bielefeld2011_Planinggruppe.JPG|270px|thumb|left|Only a well-planned project is a good project!]] | ||
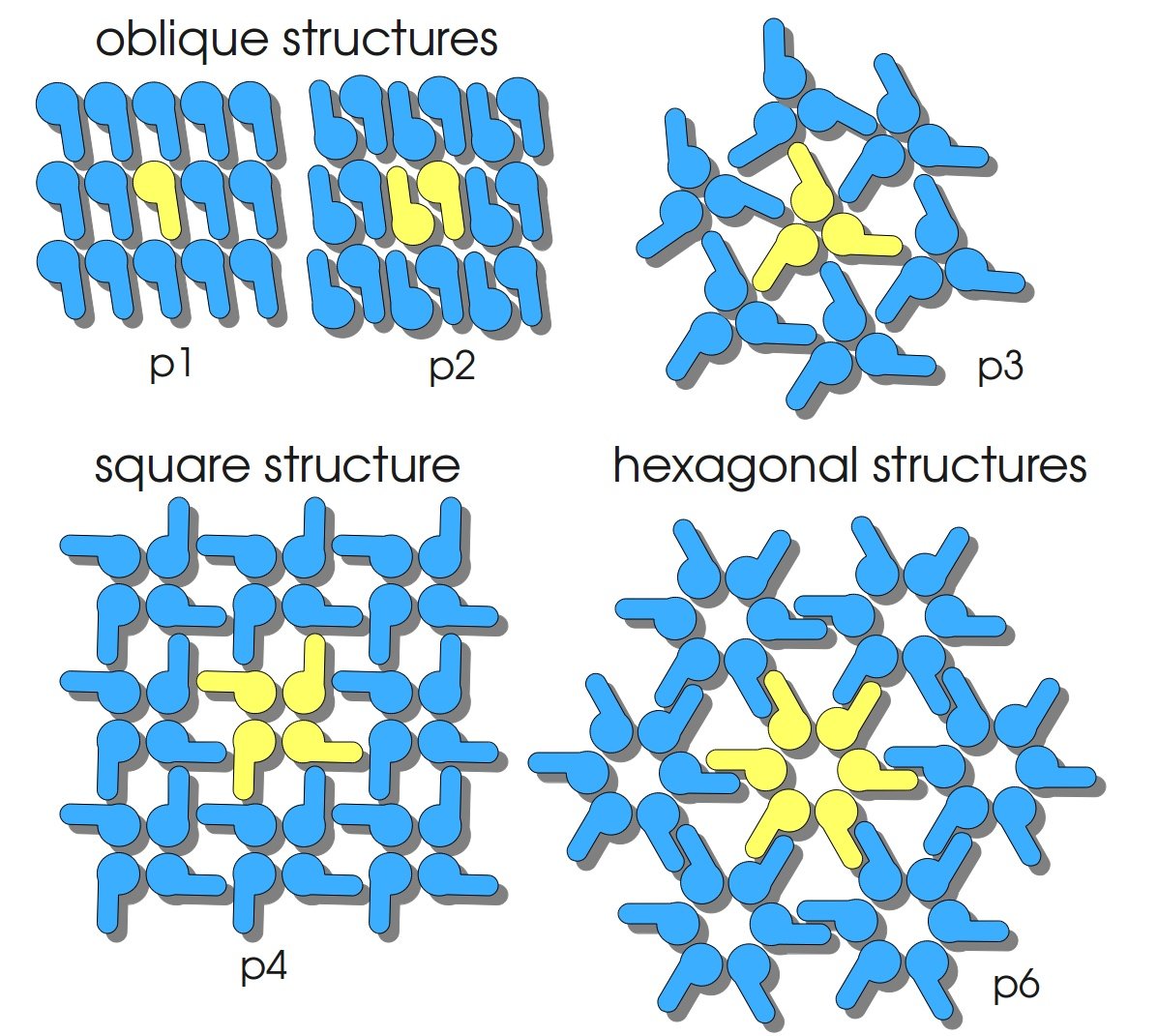
| - | + | The nanobiotechnological approach we present you here is based on so called S-layer proteins. S-layers (crystalline bacterial surface layer) are crystal-like layers consisting of multiple protein monomers and can be found in various (archae-)bacteria. They constitute the outermost part of the cell wall. Especially their ability for self-assembly into distinct geometries is of scientific interest. At phase boundaries, in solutions and on a variety of surfaces they form different lattice structures. The geometry and arrangement is determined by the C-terminal self-assembly domain, which is specific for each S-layer protein. The most common lattice geometries are oblique, square and hexagonal. By modifying the characteristics of the S-layer through combination with functional groups and protein domains as well as their defined position and orientation to each other (determined by the S-layer geometry), it is possible to realize various practical applications ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full Sleytr ''et al.'', 2007]). The usability of such well-defined nano-lattice structures is far-reaching from ultrafiltration membranes to the development of immobilized biosensors. | |
| - | <br style="clear: both" /> | + | <br style="clear:both"/> |
| - | + | [[Image:Bielefeld-Germany2011-S-Layer-Geometrien.jpg|270px|thumb|left|The most common S-layer lattice geometries.]] | |
| + | [[Image:Bielefeld-Germany-AFM.jpg|260px|thumb|left|AFM microscopy of PS2 with hexagonal lattice structure.]] | ||
So before you can build your device you have to plan it - on genetic level. We provide you two different fully BioBrick-compatible S-layer genes (<partinfo>K525301</partinfo> and <partinfo>K525401</partinfo>) which cover the lattice geometries oblique and square. Choose the one you like to use for your application, choose proteins to functionalize the nanobiotechnological surface you are going to build, model your final device (it's easy because it's cell-free so no endogenous signals and 20+ different differential equations are needed) and then: | So before you can build your device you have to plan it - on genetic level. We provide you two different fully BioBrick-compatible S-layer genes (<partinfo>K525301</partinfo> and <partinfo>K525401</partinfo>) which cover the lattice geometries oblique and square. Choose the one you like to use for your application, choose proteins to functionalize the nanobiotechnological surface you are going to build, model your final device (it's easy because it's cell-free so no endogenous signals and 20+ different differential equations are needed) and then: | ||
[[Team:Bielefeld-Germany/Results/S-Layer/Guide/2 | Let the assembly begin!]] | [[Team:Bielefeld-Germany/Results/S-Layer/Guide/2 | Let the assembly begin!]] | ||
| - | |||
Want to know more about S-layers in general? | Want to know more about S-layers in general? | ||
Latest revision as of 02:46, 29 October 2011


Planning a nanobiotechnological device
The nanobiotechnological approach we present you here is based on so called S-layer proteins. S-layers (crystalline bacterial surface layer) are crystal-like layers consisting of multiple protein monomers and can be found in various (archae-)bacteria. They constitute the outermost part of the cell wall. Especially their ability for self-assembly into distinct geometries is of scientific interest. At phase boundaries, in solutions and on a variety of surfaces they form different lattice structures. The geometry and arrangement is determined by the C-terminal self-assembly domain, which is specific for each S-layer protein. The most common lattice geometries are oblique, square and hexagonal. By modifying the characteristics of the S-layer through combination with functional groups and protein domains as well as their defined position and orientation to each other (determined by the S-layer geometry), it is possible to realize various practical applications ([http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full Sleytr et al., 2007]). The usability of such well-defined nano-lattice structures is far-reaching from ultrafiltration membranes to the development of immobilized biosensors.
So before you can build your device you have to plan it - on genetic level. We provide you two different fully BioBrick-compatible S-layer genes (<partinfo>K525301</partinfo> and <partinfo>K525401</partinfo>) which cover the lattice geometries oblique and square. Choose the one you like to use for your application, choose proteins to functionalize the nanobiotechnological surface you are going to build, model your final device (it's easy because it's cell-free so no endogenous signals and 20+ different differential equations are needed) and then:
Want to know more about S-layers in general?
Check our S-layer theory page or Sleytr UB, Huber C, Ilk N, Pum D, Schuster B, Egelseer EM (2007) S-layers as a tool kit for nanobiotechnological applications, [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6968.2006.00573.x/full FEMS Microbiol Lett 267(2):131-144].

 "
"