Team:Paris Bettencourt/Experiments/T7 diffusion experiments
From 2011.igem.org
| Line 40: | Line 40: | ||
<h2>Concentrating the cells more for microscopy (<i>B.subtilis/B.subtilis</i>)</h2> | <h2>Concentrating the cells more for microscopy (<i>B.subtilis/B.subtilis</i>)</h2> | ||
<h2>Conclusions</h2> | <h2>Conclusions</h2> | ||
| - | + | <p>During all of our experiments with the T7 polymerase diffusion we did not see any fluorescence increase (in the case of the plasmidic autoloop) nor fluorescence appearance (in the case of the chromosomic autoloop). | |
| + | We put our cells in the same conditions that we used in the <a href="https://2011.igem.org/Team:Paris_Bettencourt/GFP_diff">successful GFP diffusion experiments</a> as well as some new ones. All these repeted testing | ||
| + | gives us the only possible conclusion here: the <em>T7 RNA polymerase diffusion design show no evidence of nanotube existence so far</em>. We are currently continuing our experiments to give additional evidence to this | ||
| + | result.</p> | ||
| + | <h4>Limits</h4> | ||
| + | <p>Our plasmidic autoloop is very leaky and it is quite hard to see obvious increase in fluorescence. With even finer tuning of our experimental conditions, we might be able to see better a possible hint of nanotube | ||
| + | presence.</p> | ||
| + | <p>Our chromosomic autoloop has not been fully tested yet. We are currently trying to put it in the same cell that the T7 RNA polymerase gene to activate it all the time. The total absence of leakage under the microscope seemed quite suspect | ||
| + | at first but we saw that it could be activated in the <a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiment/T7_diff_subt_subt_microfluidic">microfluidic experiment</a>. Further investigation on this matter is under way.</p> | ||
| + | <p>We also need to properly test the T7 RNA polymerase we put in subtilis, for instance by putting a plasmidic pT7-GFP construct. However, since it is the same gene as in our pHM3 T7 autoloop, we already have indirect proof of its functionality.</p> | ||
<h2>Diffusion experiments</h2> | <h2>Diffusion experiments</h2> | ||
<h3>Results negative at first</h3> | <h3>Results negative at first</h3> | ||
Revision as of 00:34, 29 October 2011

Testing nanotubes with T7 RNA polymerase diffusion
Design overview

Schematic summary of the T7 diffusion device
All of the parts of the above design have been BioBricked, characterized both in E.coli and B.subtilis, and sent to the registry.
More details on the design are available here.
Summary
Emitter construct in E.coli - Receiver construct in B.subtilis (plasmid)
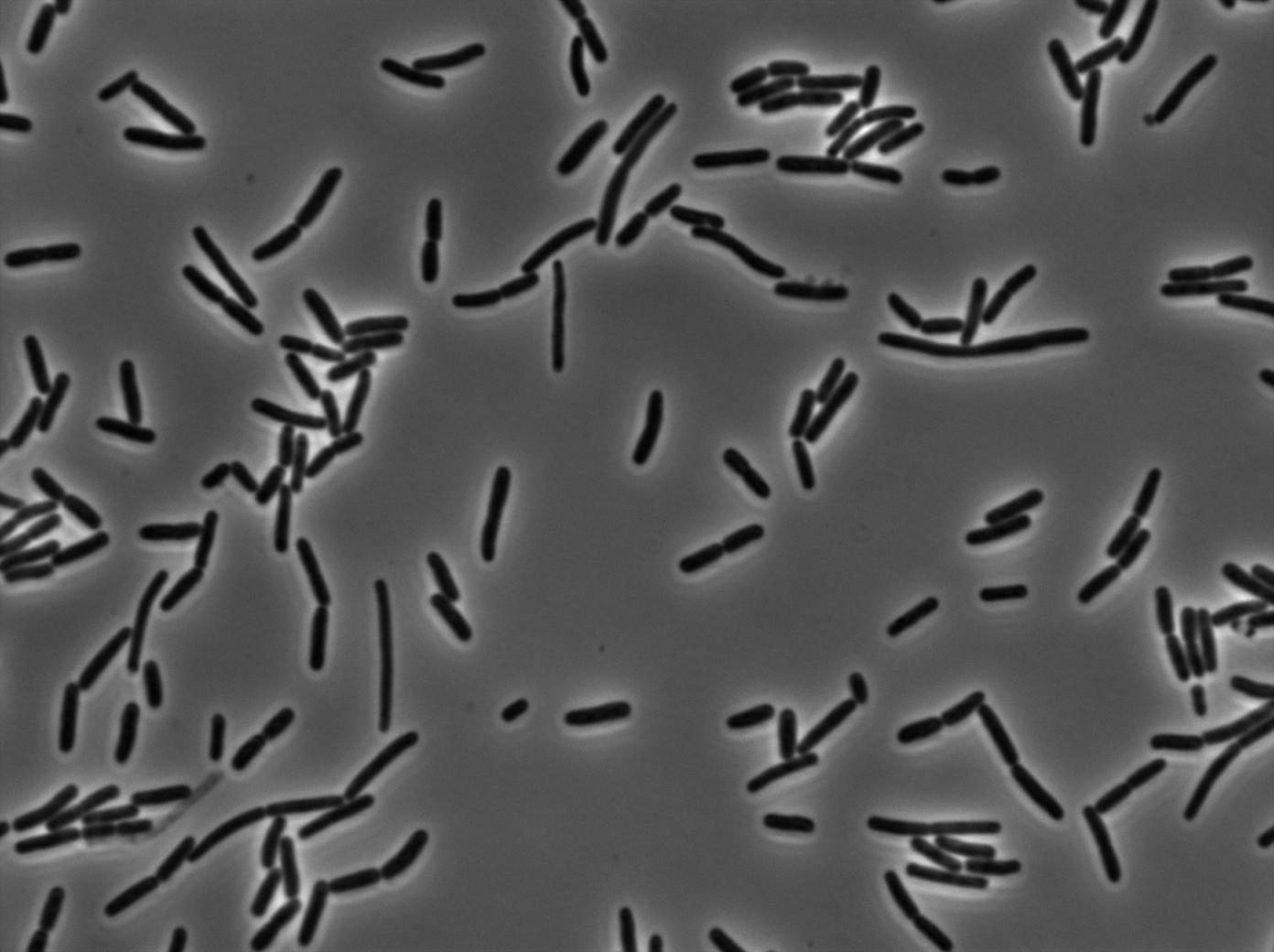
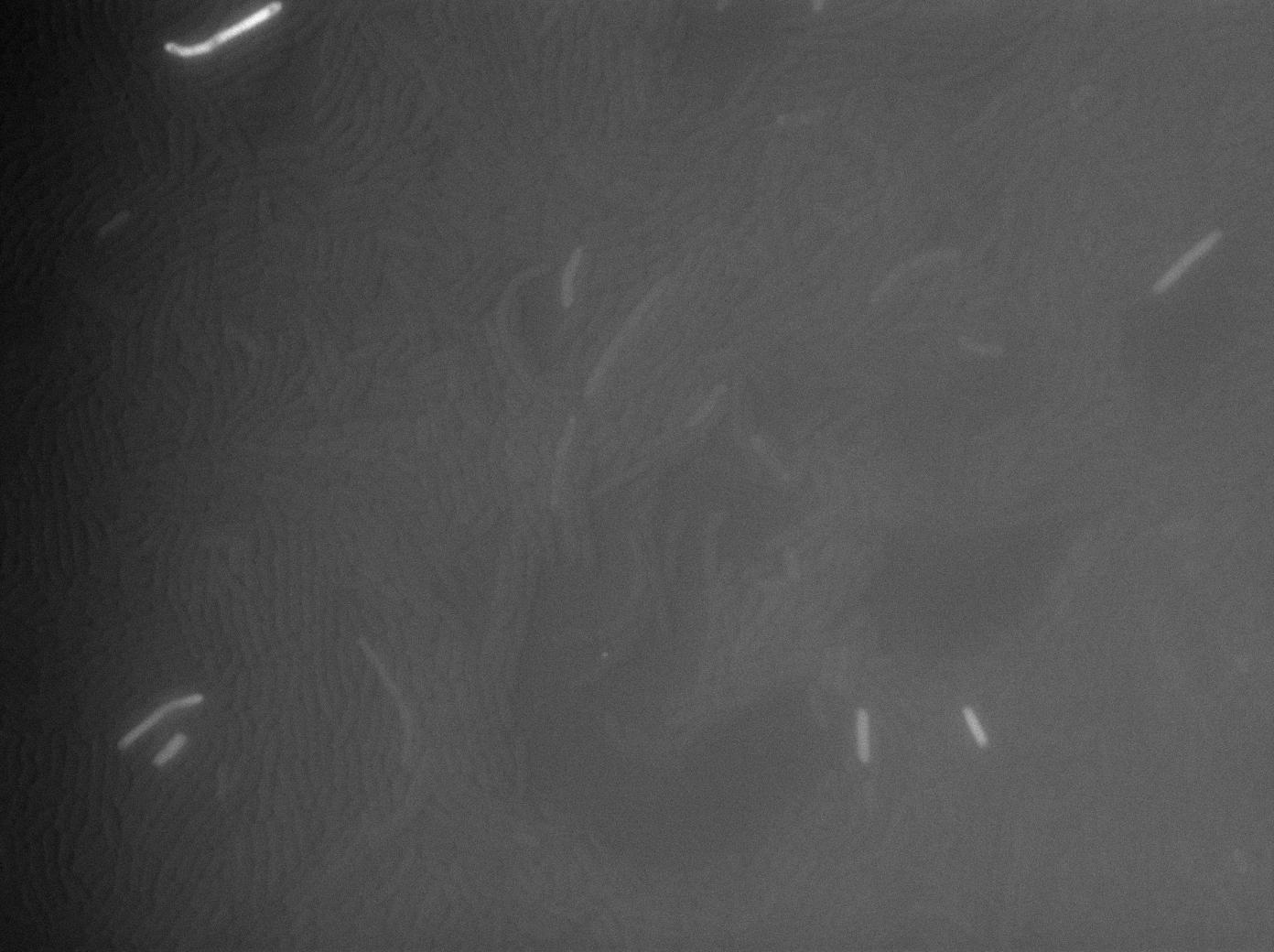
As a control for Coli to Subtillis diffusion we wanted to use E.Coli as an T7 RNA Polymerase emitter and B.Subtillis as theT7 autoloop reciever. Here we can see pictures taken using our classic microscopy protocol.As the pictures show, there is no new highly glowing cell after 125 minutes experimentation.
Emitter & receiver constructs in B.subtilis (receiver in plasmid)
Emitter & receiver constructs in B.subtilis (receiver in genome)
We used our microscopy protocol to do this experiment. The emitter strain is a B.subtilis PY9 strain where our emitter construct is integrated in the genome. The receiver strain is a B.subtilis PY9 strain where our T7 autoloop (receiver) construct is integrated in the genome.
We followed our cultures on three microscopic slides: one for the emitter, one for the receiver and one for the mix.
Using the microfluidic chip (B.subtilis/B.subtilis)
Concentrating the cells more for microscopy (B.subtilis/B.subtilis)
Conclusions
During all of our experiments with the T7 polymerase diffusion we did not see any fluorescence increase (in the case of the plasmidic autoloop) nor fluorescence appearance (in the case of the chromosomic autoloop). We put our cells in the same conditions that we used in the successful GFP diffusion experiments as well as some new ones. All these repeted testing gives us the only possible conclusion here: the T7 RNA polymerase diffusion design show no evidence of nanotube existence so far. We are currently continuing our experiments to give additional evidence to this result.
Limits
Our plasmidic autoloop is very leaky and it is quite hard to see obvious increase in fluorescence. With even finer tuning of our experimental conditions, we might be able to see better a possible hint of nanotube presence.
Our chromosomic autoloop has not been fully tested yet. We are currently trying to put it in the same cell that the T7 RNA polymerase gene to activate it all the time. The total absence of leakage under the microscope seemed quite suspect at first but we saw that it could be activated in the microfluidic experiment. Further investigation on this matter is under way.
We also need to properly test the T7 RNA polymerase we put in subtilis, for instance by putting a plasmidic pT7-GFP construct. However, since it is the same gene as in our pHM3 T7 autoloop, we already have indirect proof of its functionality.
Diffusion experiments
Results negative at first
This design was successfully implemented both in E.coli and B.subtilis. We were therefore able to test the diffusion of the T7 RNA polymerase through nanotubes. We ran several experiments:
- Diffusion from E.coli to B.subtilis with our receptor construct on a plasmid. Find more about it here
- Diffusion from B.subtilis to B.subtilis with our receptor construct on a plasmid. Find more about it here.
- Diffusion from B.subtilis to B.subtilis with our receptor construct integrated in the genome. Find more about it here.
The first three experiments gave us negative results. We saw no obvious increase of the GFP expression in receiver cells when our construct was on a plasmid and absolutely no GFP fluorescence when it was integrated in the genome.
Unexepected breakthrough
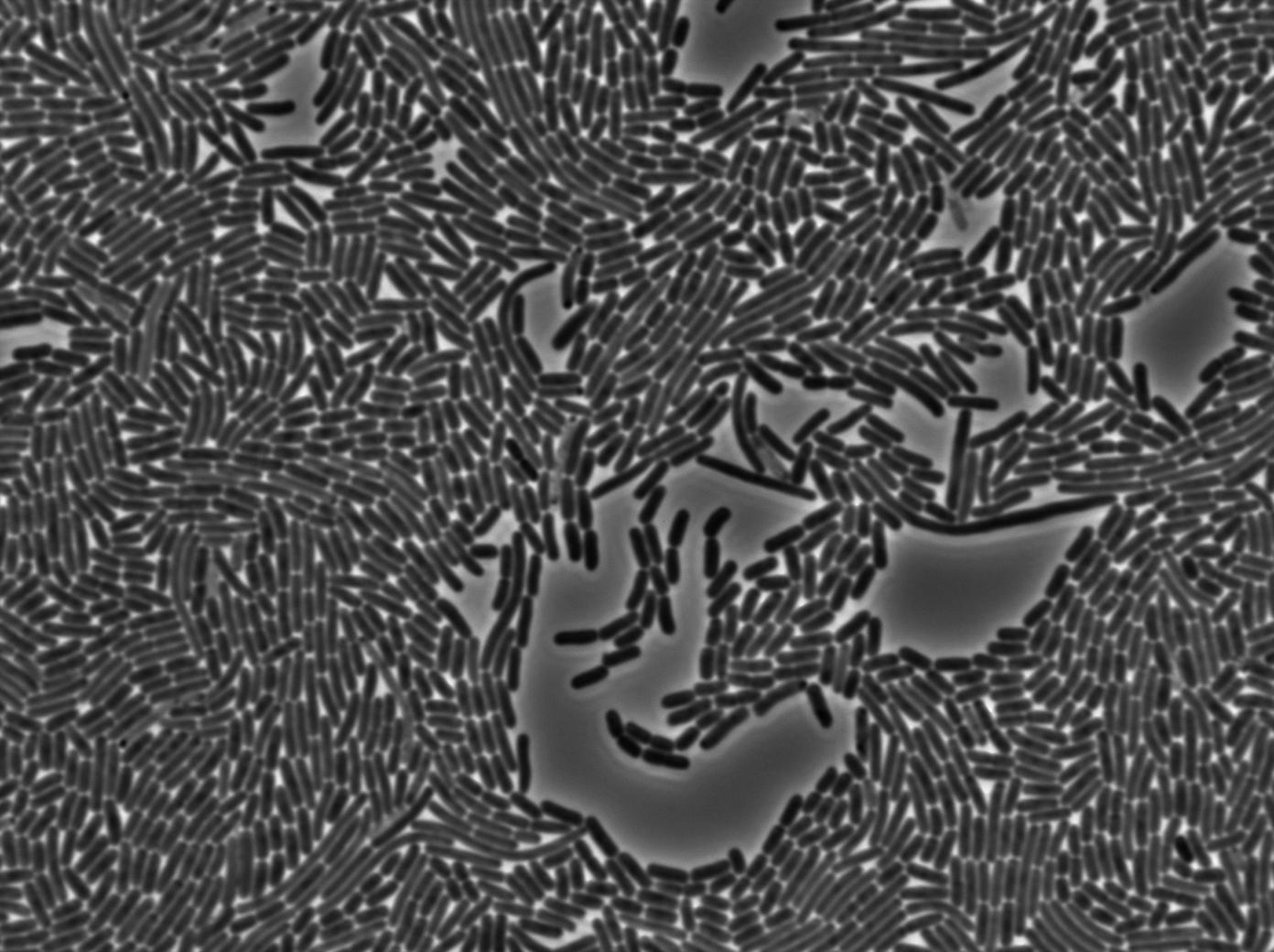
However, our microfluidic experiment gave unexepected and encouraging results. We used for this experiment two B.subtilis strains (one emitter, one receiver, both integrated in the genome). Our chromosomic T7 autoloop was brightly activated during this experiment, but only in densely packed mix of emitter and receiver cells. Find more about this experiment here.
Seeing that cell concentration seemed to be the key factor and taking advantage of our perfectly not leaky chromosomic autoloop we conducted a final set of experiments where we concentrated our cells even more. We invite you to see our final results here.
 "
"