Team:Bielefeld-Germany/Data Page
From 2011.igem.org
(Difference between revisions)
(→How our system works) |
|||
| Line 37: | Line 37: | ||
# <partinfo>K525306</partinfo> - '''Fusion protein of S-layer SgsE and mCerulean''': Characterization of induction and expression profiles of this S-layer fusion protein. | # <partinfo>K525306</partinfo> - '''Fusion protein of S-layer SgsE and mCerulean''': Characterization of induction and expression profiles of this S-layer fusion protein. | ||
# <partinfo>K525406</partinfo> - '''Fusion protein of S-layer SbpA and mCerulean''': Characterization of induction and expression profiles of this S-layer fusion protein. | # <partinfo>K525406</partinfo> - '''Fusion protein of S-layer SbpA and mCerulean''': Characterization of induction and expression profiles of this S-layer fusion protein. | ||
| + | # <partinfo>K525311</partinfo> - '''Fusion protein of S-layer SgsE and firefly luciferase''': Characterization of induction and expression profiles, purification and stability of luciferase activity of this S-layer fusion protein. | ||
Revision as of 18:21, 28 October 2011


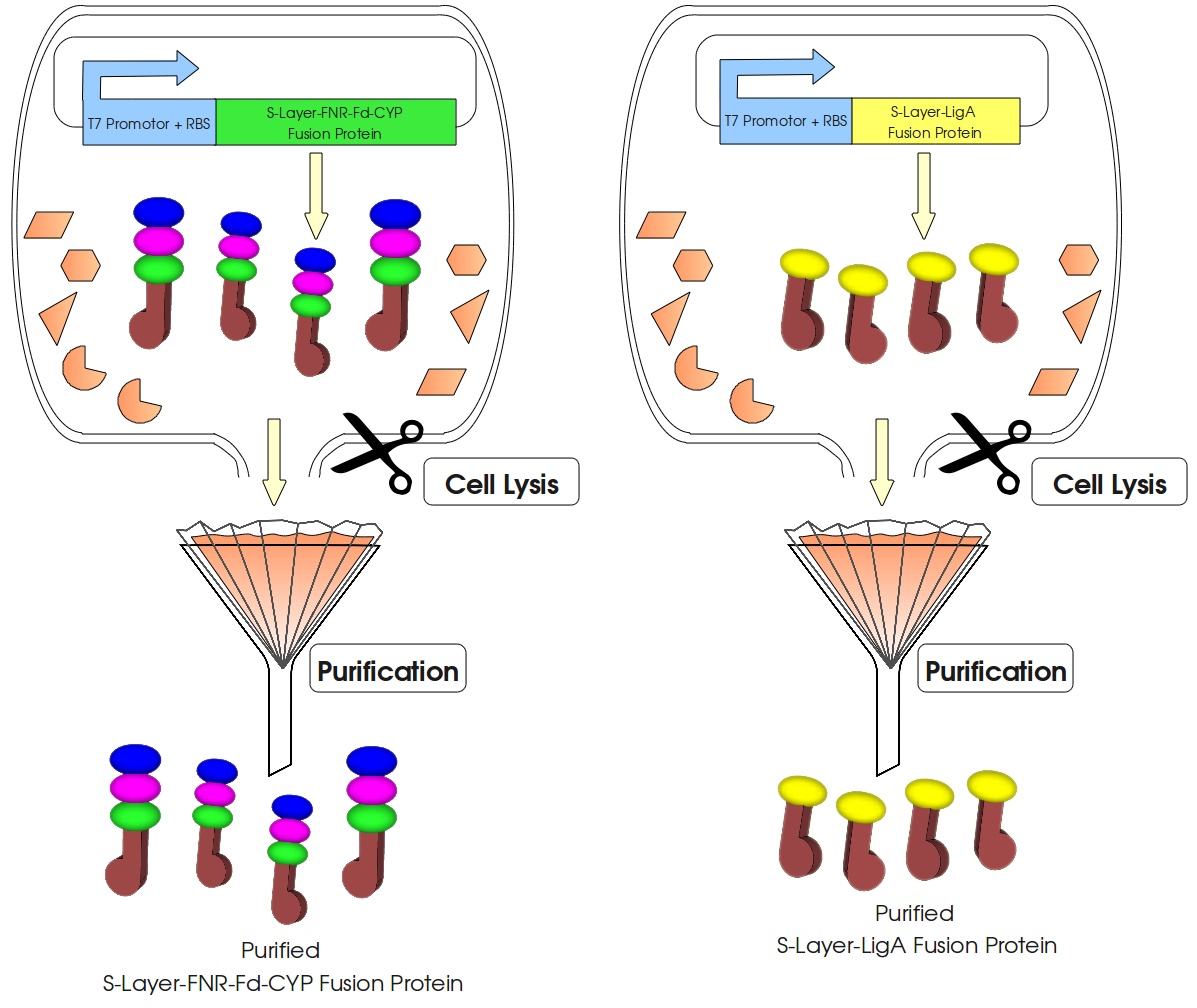
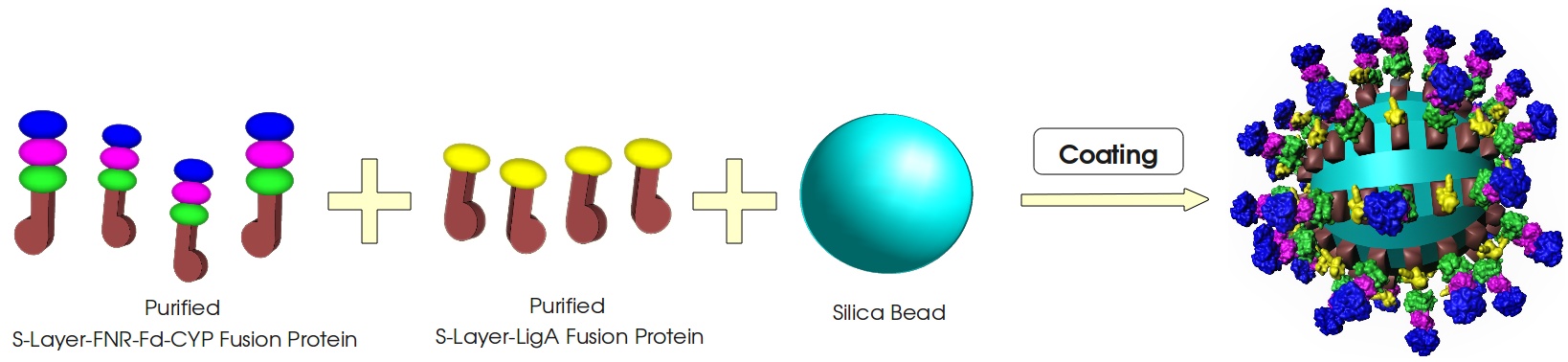
This page gives a basic overview about our cell-free bisphenol A biosensor system and the BioBricks we have used. A more detailed description of the biosensor system can be found in our project description as well as in the bisphenol A, S-layer and NAD+ detection background subsections.
Contents |
How our system works
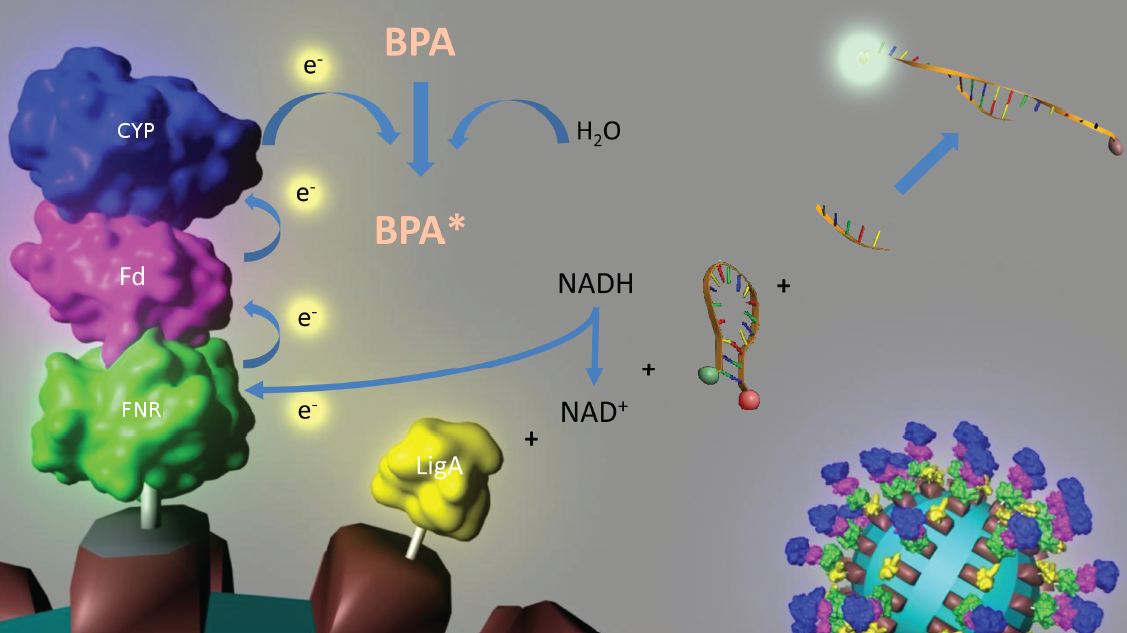
Figure 3: Visualization of our cell-free bisphenol A biosensor system with all essential components. Bisphenol A (BPA) is reduced by the electrons from NADH transferred by the ferredoxin-NADP+ oxidoreductase (FNR, <partinfo>BBa_K525499</partinfo>), ferredoxin (Fd, <partinfo>BBa_K123000</partinfo>) and cytochrome P450 (CYP, <partinfo>BBa_K123001</partinfo>) which are fused to a S-layer protein. The molecular beacon (hairpin structure) binds two short DNA oligos. The NAD+-dependent ligase (LigA, <partinfo>BBa_K525710</partinfo>), which is also fused to S-layer proteins, ligates the two oligos so that the hairpin structure opens up and the fluorophore is able to emit light after extinction.
Data for our favorite new parts
- <partinfo>K525305</partinfo> - Fusion protein of S-layer SgsE and mCitrine: This fluorescent S-layer fusion protein is used to characterize purification methods and to demonstrate the S-layers ability to self-assemble on surfaces.
- <partinfo>K525515</partinfo> - Fusion protein of BisdA and BisdB: This fusion protein improves the bisphenol A degradation in E. coli compared to the so far in the partsregistry existing BPA degrading BioBricks.
- <partinfo>K525710</partinfo> - NAD+-dependent DNA ligase from E. coli (LigA) : This enzyme enables determination of NAD+ even in very low concentrations by introducing it to a molecular beacon based bioassay.
Data for pre-existing parts
- [http://partsregistry.org/Part:BBa_K123000:Experience Experience] - BisdA degrades Bisphenol A when used with BisdB, BBa_K123000 (University of Alberta, iGEM 2008): Complete degradation of 120 mg L-1 Bisphenol A with polycistronic bisdAB gene in 30 - 33 h. Even faster (21 - 24 h) when using a fusion protein of BisdA and BisdB.
- [http://partsregistry.org/Part:BBa_K123001:Experience Experience] - BisdB degrades Bisphenol A when used with BisdA, BBa_K123001 (University of Alberta, iGEM 2008): Complete degradation of 120 mg L-1 Bisphenol A with polycistronic bisdAB gene in 30 - 33 h. Even faster (21 - 24 h) when using a fusion protein of BisdA and BisdB.
We have also characterized the following parts
- <partinfo>K525405</partinfo> - Fusion protein of S-layer SbpA and mCitrine: This fluorescent S-layer fusion protein is used to characterize purification methods and to demonstrate the S-layers ability to self-assemble on surfaces.
- <partinfo>K525512</partinfo> - Polycistronic expression of BisdA and BisdB: This is the version of BPA degrading BioBricks found in the partsregistry - comparison to our fusion protein <partinfo>K525515</partinfo>.
- <partinfo>K525517</partinfo> - Fusion Protein of BisdA and BisdB (expressed): This fusion protein improves the bisphenol A degradation in E. coli compared to the so far in the partsregistry existing BPA degrading BioBricks.
- <partinfo>K525234</partinfo> - Fusion protein of S-layer CspB and mRFP: This fluorescent S-layer fusion protein is used to characterize the intracellular localisation as well as purification methods for CspB S-layers from Corynebacterium halotolerans.
- <partinfo>K525121</partinfo> - S-layer protein CspB with TAT-sequence and lipid anchor: This fluorescent S-layer fusion protein is used to characterize the intracellular localisation and purification methods for CspB S-layers from Corynebacterium glutamicum.
- <partinfo>K525123</partinfo> - S-layer protein CspB with lipid anchor: This fluorescent S-layer fusion protein is used to characterize the intracellular localisation and purification methods for CspB S-layers from Corynebacterium glutamicum.
- <partinfo>K525222</partinfo> - S-layer protein CspB: This fluorescent S-layer fusion protein is used to characterize the intracellular localisation and purification methods for CspB S-layers from Corynebacterium halotolerans.
- <partinfo>BBa_K525223</partinfo> - S-layer protein CspB with lipid anchor: This fluorescent S-layer fusion protein is used to characterize the intracellular localisation and purification methods for CspB S-layers from Corynebacterium halotolerans.
- <partinfo>K525224</partinfo> - S-layer protein CspB with TAT-sequence: This fluorescent S-layer fusion protein is used to characterize the intracellular localisation and purification methods for CspB S-layers from Corynebacterium halotolerans.
- <partinfo>K525304</partinfo> - Fusion protein of S-layer SbpA and mCherry: Characterization of induction and expression profiles of this S-layer fusion protein.
- <partinfo>K525306</partinfo> - Fusion protein of S-layer SgsE and mCerulean: Characterization of induction and expression profiles of this S-layer fusion protein.
- <partinfo>K525406</partinfo> - Fusion protein of S-layer SbpA and mCerulean: Characterization of induction and expression profiles of this S-layer fusion protein.
- <partinfo>K525311</partinfo> - Fusion protein of S-layer SgsE and firefly luciferase: Characterization of induction and expression profiles, purification and stability of luciferase activity of this S-layer fusion protein.
 "
"