Team:Harvard/Results/Biobricks
From 2011.igem.org
(→TolC one-hybrid system selection strain (BBa_K615003)) |
(→His3-URA3 One-hybrid selection system strain (BBa_K61500)) |
||
| (13 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{:Team:Harvard/Template:ResultsBar}} | {{:Team:Harvard/Template:ResultsBar}} | ||
{{:Team:Harvard/Template:ResultsGrayBar}} | {{:Team:Harvard/Template:ResultsGrayBar}} | ||
| + | <div class="whitebox"> | ||
| + | See especially our favorites: [https://2011.igem.org/Team:Harvard/Results/Biobricks#His3-URA3_One-hybrid_selection_system_strain_.28BBa_K61500.29 BBa_K615000], [https://2011.igem.org/Team:Harvard/Results/Biobricks#TolC_one-hybrid_system_selection_strain_.28BBa_K615003.29 BBa_K615003], and [https://2011.igem.org/Team:Harvard/Results/Biobricks#Zif268_binding_site-His3-URA3_construct_for_one-hybrid_selection_.28BBa_K615004.29 BBa_K615004]. | ||
| + | </div> | ||
<div class="whitebox"> | <div class="whitebox"> | ||
__toc__ | __toc__ | ||
| Line 9: | Line 12: | ||
[http://partsregistry.org/Part:BBa_K615000 See our chassis page] in the Registry of Standard Biological Parts. | [http://partsregistry.org/Part:BBa_K615000 See our chassis page] in the Registry of Standard Biological Parts. | ||
| - | The His3-URA3 one-hybrid selection system has been developed by Meng et al (2005)to test zinc finger binding by tying the binding event to cell survival. However, Meng used a plasmid-based system where both the zinc fingers and the binding site were on vectors within the cell, while we took advantage of recent advances in technology to make this one-hybrid system genome based. This prevents one or both components from being lost during cell division, ensures that each cell has exactly one copy of the zinc finger binding site-His3-URA3 construct, and presents the genome as the next frontier for standardized part construction. (For a full description of the one-hybrid system, see our [https://2011.igem.org/Team:Harvard/Project/ | + | The His3-URA3 one-hybrid selection system has been developed by Meng et al (2005)to test zinc finger binding by tying the binding event to cell survival. However, Meng used a plasmid-based system where both the zinc fingers and the binding site were on vectors within the cell, while we took advantage of recent advances in technology to make this one-hybrid system genome based. This prevents one or both components from being lost during cell division, ensures that each cell has exactly one copy of the zinc finger binding site-His3-URA3 construct, and presents the genome as the next frontier for standardized part construction. (For a full description of the one-hybrid system, see our [https://2011.igem.org/Team:Harvard/Project/Test Test page].) |
| - | Using lambda red and MAGE, we knocked out HisB and PyrF and replaced with them with the yeast analogs His3 and URA3 under the control of a zinc finger binding site (see our [https://2011.igem.org/Team:Harvard/ | + | Using lambda red and MAGE, we knocked out HisB and PyrF and replaced with them with the yeast analogs His3 and URA3 under the control of a zinc finger binding site (see our [https://2011.igem.org/Team:Harvard/Test Test] page). The omega subunit of RNA polymerase was also knocked out and instead fused to the first zinc finger of the array to be tested (see below). This allows zinc finger binding to control expression of His3 and URA3, and thus in incomplete media the cell will only survive if the proper zinc finger is present. |
| - | Our strain was carefully characterized and shown to have the proper phenotype and be sensitive enough to recognize a valid zinc finger when diluted up to one in one million. The selectivity can also be adjusted by changing the concentration of 3-AT, a competitive inhibitor of His3 (see [https://2011.igem.org/Team:Harvard/ | + | Our strain was carefully characterized and shown to have the proper phenotype and be sensitive enough to recognize a valid zinc finger when diluted up to one in one million. The selectivity can also be adjusted by changing the concentration of 3-AT, a competitive inhibitor of His3 (see [https://2011.igem.org/Team:Harvard/Project/Test#Fine-tuning_the_one-hybrid_selection_system Fine Tuning One Hybrid Selection] for details). See below for a summary of the changes made to the strain and how to successfully use it. |
| Line 33: | Line 36: | ||
Courtesy of the Church Lab. | Courtesy of the Church Lab. | ||
| + | |||
| + | =Zif268 binding site-His3-URA3 construct for one-hybrid selection (BBa_K615004)= | ||
| + | [http://partsregistry.org/Part:BBa_K615004 See the Biobrick page] in the Registry of Standard Biological Parts. | ||
| + | |||
| + | This biobrick contains the Zif268 binding site-His3-URA3 construct needed for the His3-URA3 one hybrid selection system. This can be used to build one's own genome-based strain by attaching the construct to an antibiotic resistance cassette and using lambda red to insert into the genome (as the our team did), or it can be used as is, as part of a plasmid-based selection system. Besides characterizing the genomic version of the selection system (see above), we also used this construct as plasmid expressed in a HisB, PyrF, rpoZ KO strain. We found that it produced identical growth curves to the genome-based version, as seen in the example below: | ||
| + | |||
| + | |||
| + | [[File: HARVzfb1.png|center]] | ||
=Zinc Finger-Omega Subunit Fusion Protein (BBa_K615001)= | =Zinc Finger-Omega Subunit Fusion Protein (BBa_K615001)= | ||
| Line 41: | Line 52: | ||
This specific part is designed to bind to the sequence 5'-GTG GGA TGG-3', where the GTG-GGA codons have known zinc finger binders, but the GGA-TGG (F2-F1) combination was unknown. Finding a zinc finger that binds to TGG in the context of GGA can also translate into many three-finger arrays, since any known F3-GGA pair can be combined with F2 GGA-F1 TGG. | This specific part is designed to bind to the sequence 5'-GTG GGA TGG-3', where the GTG-GGA codons have known zinc finger binders, but the GGA-TGG (F2-F1) combination was unknown. Finding a zinc finger that binds to TGG in the context of GGA can also translate into many three-finger arrays, since any known F3-GGA pair can be combined with F2 GGA-F1 TGG. | ||
| - | [[File: HARVbb001_2.png]] | + | [[File: HARVbb001_2.png|center]] |
</div> | </div> | ||
<div class="whitebox"> | <div class="whitebox"> | ||
| + | |||
=References= | =References= | ||
Xiangdong Meng, Michael H Brodsky, Scot A Wolfe. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. (2005) Nature Biotechnology, 23(8): 988-994. | Xiangdong Meng, Michael H Brodsky, Scot A Wolfe. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. (2005) Nature Biotechnology, 23(8): 988-994. | ||
Latest revision as of 23:48, 25 October 2011
Overview | Biobricks | Source Code | Acknowledgments | Accomplishments
See especially our favorites: BBa_K615000, BBa_K615003, and BBa_K615004.
Contents |
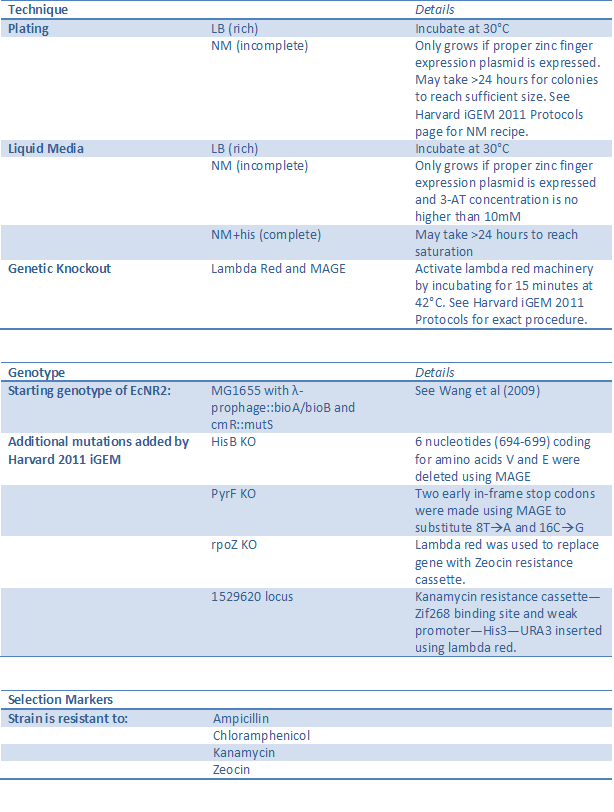
His3-URA3 One-hybrid selection system strain (BBa_K61500)
[http://partsregistry.org/Part:BBa_K615000 See our chassis page] in the Registry of Standard Biological Parts.
The His3-URA3 one-hybrid selection system has been developed by Meng et al (2005)to test zinc finger binding by tying the binding event to cell survival. However, Meng used a plasmid-based system where both the zinc fingers and the binding site were on vectors within the cell, while we took advantage of recent advances in technology to make this one-hybrid system genome based. This prevents one or both components from being lost during cell division, ensures that each cell has exactly one copy of the zinc finger binding site-His3-URA3 construct, and presents the genome as the next frontier for standardized part construction. (For a full description of the one-hybrid system, see our Test page.)
Using lambda red and MAGE, we knocked out HisB and PyrF and replaced with them with the yeast analogs His3 and URA3 under the control of a zinc finger binding site (see our Test page). The omega subunit of RNA polymerase was also knocked out and instead fused to the first zinc finger of the array to be tested (see below). This allows zinc finger binding to control expression of His3 and URA3, and thus in incomplete media the cell will only survive if the proper zinc finger is present.
Our strain was carefully characterized and shown to have the proper phenotype and be sensitive enough to recognize a valid zinc finger when diluted up to one in one million. The selectivity can also be adjusted by changing the concentration of 3-AT, a competitive inhibitor of His3 (see Fine Tuning One Hybrid Selection for details). See below for a summary of the changes made to the strain and how to successfully use it.
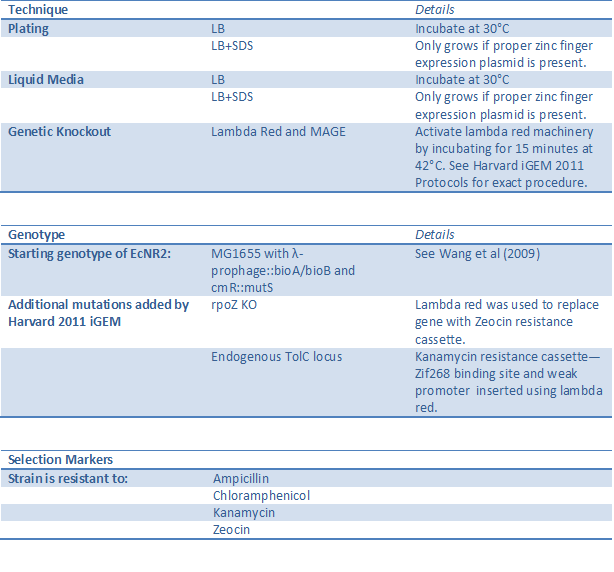
TolC one-hybrid system selection strain (BBa_K615003)
[http://partsregistry.org/Part:BBa_K615003 See our chassis page] on the Registry of Standard Biological Parts.
Similar to the His3-URA3 one-hybrid system, the TolC strain uses a one-hybrid selection system to test for zinc finger binding. Although less sensitive than the His3-URA3 system (can only distinguish a valid hit in a dilution of one to one hundred), TolC has both the positive and the negative selection components embodied in the same gene. TolC is a membrane pump needed to remove toxins from the cell, so when the strain is grown in media containing SDS, cells will only survive if the proper zinc finger-omega subunit fusion protein is bound upstream of the endogenous TolC gene. To eliminate clones with leaky promoters, the cells can be grown in media containing colicin, a toxin that requires TolC to enter the cell. See below for a summary of features and techniques that can be used with this strain.
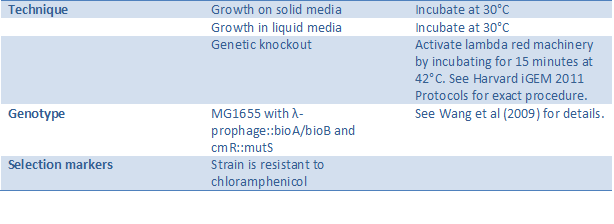
EcNR2 strain (BBa_K615002)
[http://partsregistry.org/Part:BBa_K615002 See our chassis page] in the Registry of Standard Biological Parts.
EcNR2 is a modified strain of E. coli designed for easy manipulation of the genome (see Wang et al). This strain is the ideal chassis for projects involving MAGE and lambda red, since it contains the lambda red machinery and MutS knockout needed to make such genome editing efficient. Our one-hybrid selection systems were all created using EcNR2, proving that both techniques work successfully in this strain. See below for a summary of features and techniques.
Courtesy of the Church Lab.
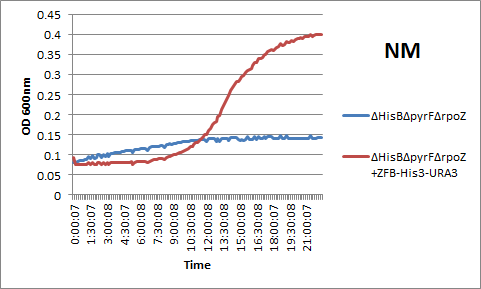
Zif268 binding site-His3-URA3 construct for one-hybrid selection (BBa_K615004)
[http://partsregistry.org/Part:BBa_K615004 See the Biobrick page] in the Registry of Standard Biological Parts.
This biobrick contains the Zif268 binding site-His3-URA3 construct needed for the His3-URA3 one hybrid selection system. This can be used to build one's own genome-based strain by attaching the construct to an antibiotic resistance cassette and using lambda red to insert into the genome (as the our team did), or it can be used as is, as part of a plasmid-based selection system. Besides characterizing the genomic version of the selection system (see above), we also used this construct as plasmid expressed in a HisB, PyrF, rpoZ KO strain. We found that it produced identical growth curves to the genome-based version, as seen in the example below:
Zinc Finger-Omega Subunit Fusion Protein (BBa_K615001)
[http://partsregistry.org/Part:BBa_K615001 See the Biobrick page] in the Registry of Standard Biological Parts.
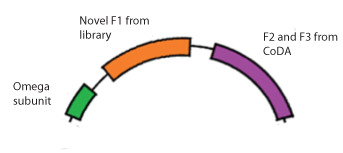
To test our library of 55,000 zinc fingers, we designed expression plasmids that contain the second and third zinc fingers (F2 and F3) in the array that are known from CoDA, the novel finger from the library, and the omega subunit. When transformed into the selection strain, the ability of the zinc finger to bind will determine whether RNA polymerase is recruited and thus whether His3 and URA3 are expressed. (See our Chip Library page for construction details.)
This specific part is designed to bind to the sequence 5'-GTG GGA TGG-3', where the GTG-GGA codons have known zinc finger binders, but the GGA-TGG (F2-F1) combination was unknown. Finding a zinc finger that binds to TGG in the context of GGA can also translate into many three-finger arrays, since any known F3-GGA pair can be combined with F2 GGA-F1 TGG.
References
Xiangdong Meng, Michael H Brodsky, Scot A Wolfe. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. (2005) Nature Biotechnology, 23(8): 988-994.
Jeffry D Sander, Elizabeth J Dahlborg, Mathew J Goodwin et al. Selection-free zinc-finger-nuclease engineering by contextdependent assembly (CoDA). (2011) Nature Methods 8(1): 53-55.
Harris H. Wang, Farren J. Isaacs, Peter A. Carr, Zachary Z. Sun, George Xu, Craig R. Forest, George M. Church. Programming cells by multiplex genome engineering and accelerated evolution. (2009). Nature, 460(7257):894-8.
 "
"