Team:Washington/Outreach/iGEM Collaborations
From 2011.igem.org
(Difference between revisions)
(→Software Testing) |
|||
| (16 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
| + | <center><big><big><big><big>'''Community Outreach: iGEM Collaborations'''</big></big></big></big></center><br><br> | ||
| - | =Primer Design Tool with LMU-Munich= | + | ='''Primer Design Tool with LMU-Munich'''= |
| - | We worked with [https://2011.igem.org/Team:LMU-Munich LMU-Munich 2011] iGEM team this summer on the testing and development of their PrimerDesign tool. To learn more about it check out their | + | We worked with [https://2011.igem.org/Team:LMU-Munich LMU-Munich 2011] iGEM team this summer on the testing and development of their PrimerDesign tool. To learn more about it check out their [https://2011.igem.org/Team:LMU-Munich/Primer_Design/Design_Primers website]. |
| - | |||
| - | + | ---- | |
| - | [[ | + | |
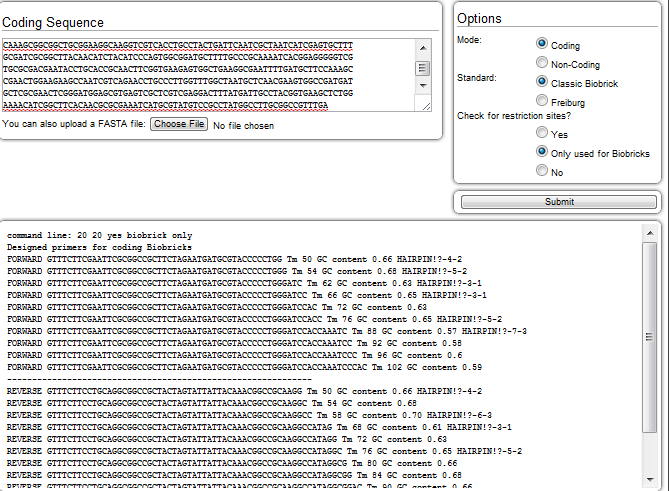
| + | [[Image:Washington_collab_test.png|right|550px|thumb|An example of the output from the tool based off of input from our luxC biobrick, multiple primers with various melting temperatures are given]] | ||
| + | |||
| + | =='''Software Testing'''== | ||
| + | |||
| + | We helped LMU-Munich test their primer design software by sending our primer data and comparing it with the output of their automated designer. To do this, we compared the sequences of the primers used to amplify out parts of the LuxBrick with the primers that their software designed and concluded that their software works as expected. Unfortunately, we did not have enough time to order the primers that the software designed to test it ourselves. Overall, we found the tool easy to use and potentially very useful for future primer design. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =='''Implemented Bug Fixes & Features'''== | ||
| + | #Added cyanobacteria to available genomes | ||
| + | #Switch coding sequences in the primer designer to allow for the user to incude a stop codon in a coding sequence. Before, software insisted that coding sequences not include the stop codon | ||
| + | #Made primer designer default to only check for biobrick restriction enzymes. Before, Primer Designer defaulted to checking for all restriction sites, making the user scroll to the bottom of the page for results | ||
| + | #Added support for FASTA files with headers | ||
| + | #Added descriptions for each column of the Primer Designer output. | ||
| + | |||
| + | |||
| + | <center><gallery caption="Original vs. Revised Output" widths="425px" heights="250px" perrow="2"> | ||
| + | Image:Washington_OldPrimerDesignerdefault.png|<center>'''Original Output'''</center><br> The original output identified every commercially available enzyme recognition site by default. This made it annoying to reach the bottom of the page where the primer info was. | ||
| + | Image:Washington_primernewdefault.png|<center>'''Revised Output'''</center> <br> New, more useful default output with only searching for BioBrick enzymes, and providing labels in the output | ||
| + | </gallery></center> | ||
| + | |||
| + | <gallery> | ||
| + | [[File:Washington_OldPrimerDesignerdefault.png|left|550px|thumb|Old Default Primer Designer Output]] | ||
| + | [[File:Washington_primernewdefault.png|left|550px|thumb|New, more useful default output with only searching for BioBrick enzymes, and providing labels in the output]] | ||
| + | <gallery/> | ||
Latest revision as of 20:31, 23 September 2011
Primer Design Tool with LMU-Munich
We worked with LMU-Munich 2011 iGEM team this summer on the testing and development of their PrimerDesign tool. To learn more about it check out their website.
Software Testing
We helped LMU-Munich test their primer design software by sending our primer data and comparing it with the output of their automated designer. To do this, we compared the sequences of the primers used to amplify out parts of the LuxBrick with the primers that their software designed and concluded that their software works as expected. Unfortunately, we did not have enough time to order the primers that the software designed to test it ourselves. Overall, we found the tool easy to use and potentially very useful for future primer design.
Implemented Bug Fixes & Features
- Added cyanobacteria to available genomes
- Switch coding sequences in the primer designer to allow for the user to incude a stop codon in a coding sequence. Before, software insisted that coding sequences not include the stop codon
- Made primer designer default to only check for biobrick restriction enzymes. Before, Primer Designer defaulted to checking for all restriction sites, making the user scroll to the bottom of the page for results
- Added support for FASTA files with headers
- Added descriptions for each column of the Primer Designer output.
 "
"