Team:ETH Zurich/Biology/Validation
From 2011.igem.org
(→Results) |
|||
| Line 31: | Line 31: | ||
== Results == | == Results == | ||
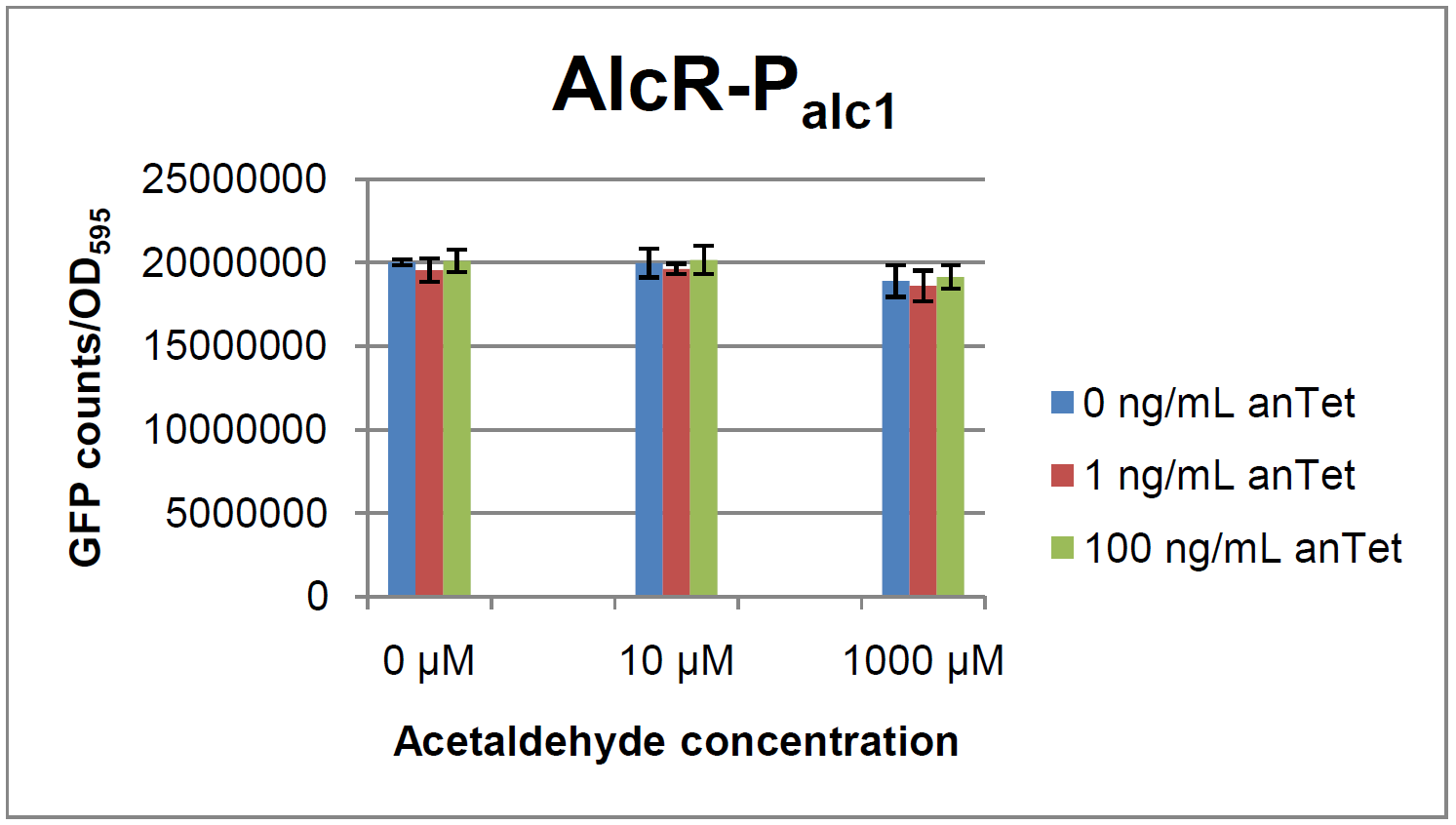
| - | + | In a first experiment, the ''alc''-promoters were tested by using a non-codon optimized natural variant of ''alcR''. Because of this we tested the compatibility of the codon usage in the variant from ''Aspergillus nidulans'' with the one from ''E. coli''. We received a codon adaptation index (CAI) of 0.7 [[#Ref2[2]]]. Nevertheless, some rare codons in ''E.coli'' were present in the first 20 amino acids of the gene. After a first experiment without any significant results (data not shown), we therefore optimized the 20 first amino acids of ''alcR'' by PCR and also ordered a fully codon-optimized variant. All of the following experiments were performed with ''alcR'' codon-optimized within the first 20 amino acids. | |
[[File:Test Palc1.png|400px|left|thumb|'''Test results for P<sub>alc1</sub>''']] | [[File:Test Palc1.png|400px|left|thumb|'''Test results for P<sub>alc1</sub>''']] | ||
Revision as of 12:01, 21 September 2011
| Validation |
| ||||
| Experimental setups... | |||||
AlcR sensorExperimental setupAs described in the Smoke detectors section we designed two artificial AlcR-dependent promoters which get repressed upon binding of AlcR to acetaldehyde. For testing of our design we created a system with a superfolded GFP output in order to characterize the gene expression with different levels of AlcR production and acetaldehyde induction. The system work the following way: A) In normal medium conditions the TetR transcription factor is produced and binds to its cognate promoter, thus inhibiting the expression of alcR. In this case no AlcR is present and GFP is produced. B) Upon addition of increasing amounts of anhydrotetracycline, TetR gets released from the promoter and AlcR is produced. The amount of AlcR should increase with increasing amounts of anhydrotetracycline. C) By addition of acetaldehyde AlcR binds to the alc-promoter and the GFP response decreases.
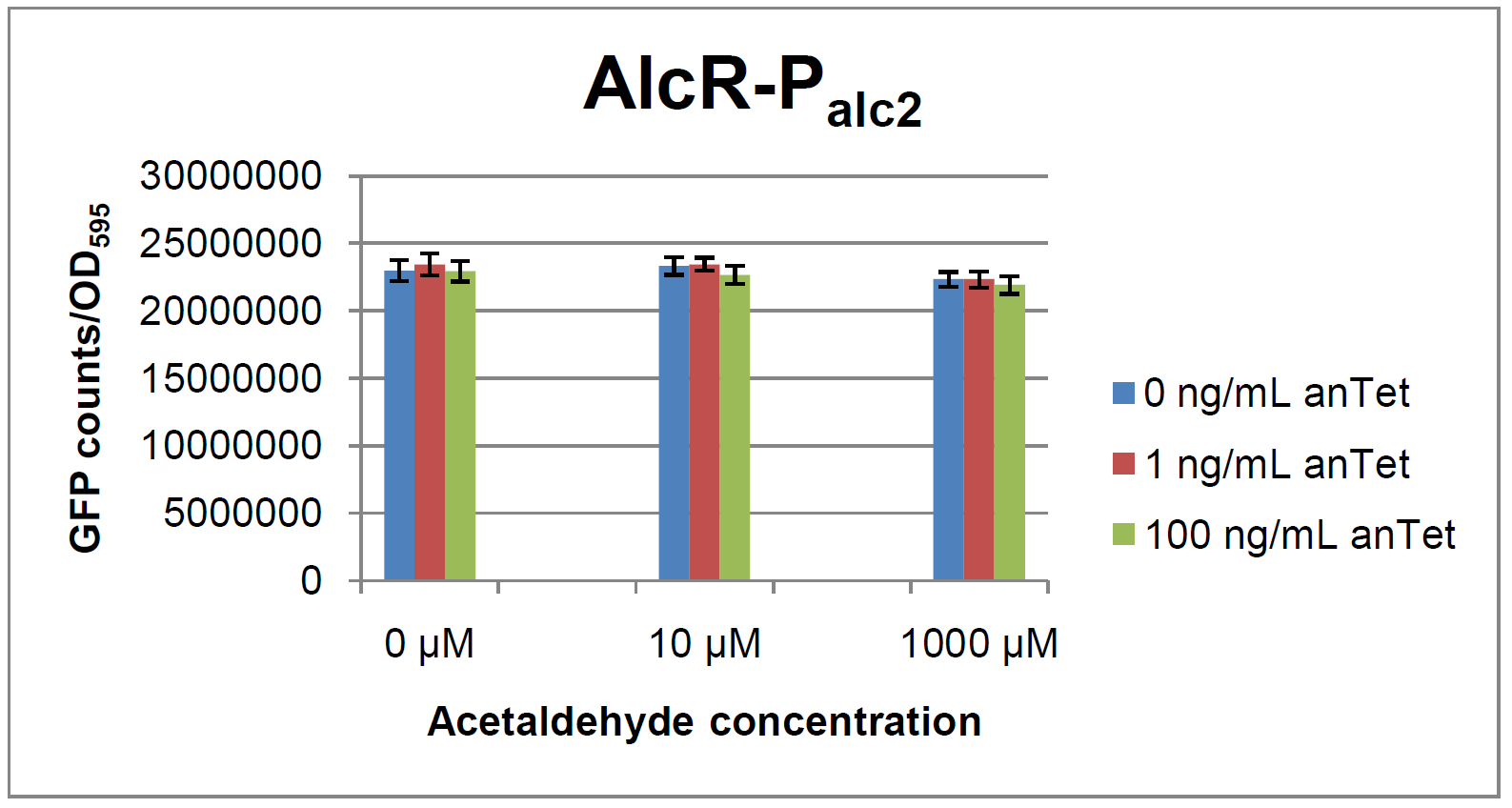
ResultsIn a first experiment, the alc-promoters were tested by using a non-codon optimized natural variant of alcR. Because of this we tested the compatibility of the codon usage in the variant from Aspergillus nidulans with the one from E. coli. We received a codon adaptation index (CAI) of 0.7 [[#Ref2[2]]]. Nevertheless, some rare codons in E.coli were present in the first 20 amino acids of the gene. After a first experiment without any significant results (data not shown), we therefore optimized the 20 first amino acids of alcR by PCR and also ordered a fully codon-optimized variant. All of the following experiments were performed with alcR codon-optimized within the first 20 amino acids. Expression of AlcR in E. coliGiven that codon usage is not the same in E. coli and Aspergillus nidulans, we analyzed codon usage of AlcR. We obtained a Codon adaptation index (CAI) of 0.7 for the Aspergillus nidulans protein in E. coli, especially at the beginning of the gene rare codons were included. To get a fully E. coli-optimized gene, we ordered the gene codon optimized. To make some first test, we exchanged the rare codons at the beginning of the Aspergillus nidulans protein with PCR. With these protein we made same first Test of the expression of AlcR. To monitor the expression of AlcR a His-tag was introduced in the test system. western blot Repression test of AlcRTo test our designed AlcR-promotors the following system was designed (see Figure). In present of acetaldeyhde AlcR binds to the AlcR operon and inhibits the expression of GFP. For a better signal assam gfp was used. results
|
XylR sensor |
Xylene degradation |
Bandpass |
References[1] [http://www.ncbi.nlm.nih.gov/pubmed/11550794 Beatrice Felenbok, Michel Flipphi and Igor Nikolaev: Ethanol Catabolism in Aspergillus nidulans: A Model System for Studying Gene Regulation, Progress in Nucleic Acid Research and Molecular Biology, 69: 149-204] |
 "
"