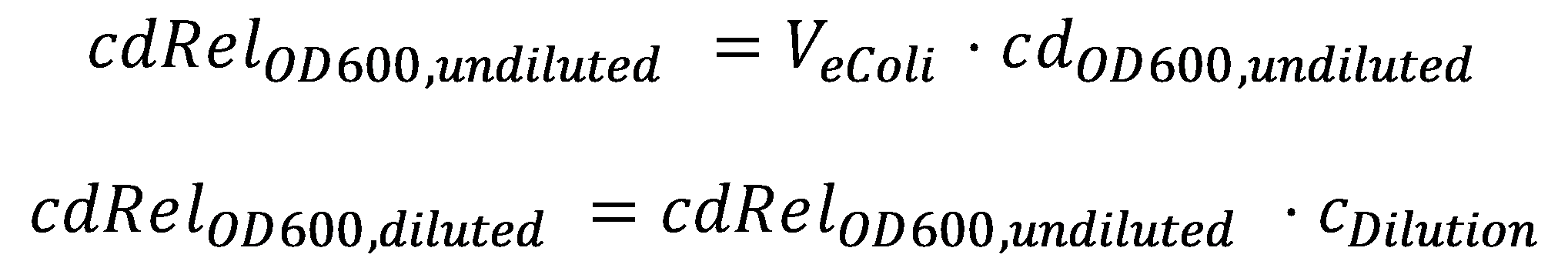Team:ETH Zurich/Modeling/Microfluidics
From 2011.igem.org
(→Diffusion) |
(→Diffusion) |
||
| Line 26: | Line 26: | ||
[[File:ETHZ-Diffusion.png|196px|center]] | [[File:ETHZ-Diffusion.png|196px|center]] | ||
| + | For the relevant diffusion parameter, D<sub>AcAl</sub>, see [[Team:ETH_Zurich/Modeling/Parameters|the parameters page]]. | ||
|} | |} | ||
{|class="roundContainer" | {|class="roundContainer" | ||
Revision as of 15:09, 16 September 2011
| Microfluidics Model |
| ||||
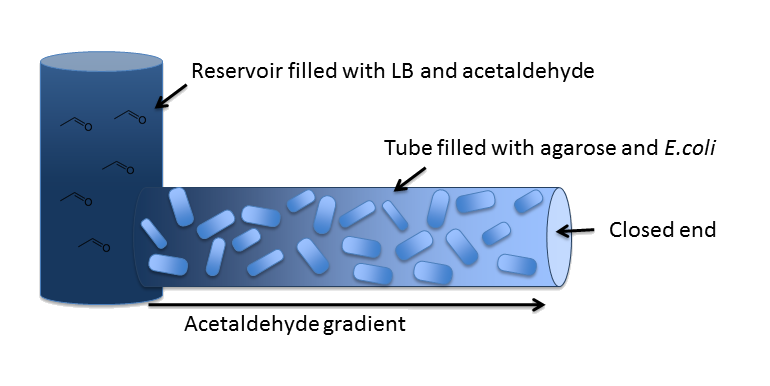
| In our first spatiotemporal model, we wanted to find out if an Acetaldehyde gradient would be formed at all. The idea was to generate it as an equilibrium of Acetaldehyde diffusion out of the reservoir into our channel and Acetaldehyde degradation by the cells in it. In order to answer this question, we modeled a spatiotemporal reaction-diffusion system, first in 2D with MATLAB and then in 3D with COMSOL Multiphysics. | |||||
Overview |
DiffusionFirst of all, we have to model diffusion of Acetaldehyde from the reservoir to the closed end of the microfluidics channel. The relevant equation is the one for isotropic diffusion, i.e. diffusion that is uniform in all directions: For the relevant diffusion parameter, DAcAl, see the parameters page. |
DegradationCell DensityIn order to able to determine how much Acetaldehyde our smoColi cells are able to degrade, we first need to determine the cell density of the immobilized cells in agarose. For this calculation, we assume we have a cell culture with OD600 with which we fill the channel. Parameter-wise, to determine the relative cell density in the channel voume, we need to know:
A parameter we can choose suitably is the dilution of the cells in agarose, cDilution. Then we can calculate the relative cell density in the smoColi-agarose-filled microfluidics channel: |
Simulation |
 "
"