Team:Freiburg/Notebook/5 September
From 2011.igem.org

Contents |
green light receptor
transformation with reporter parts
Investigators:Julia
transformation with iGEM parts [http://partsregistry.org/wiki/index.php?title=Part:BBa_I15016 BBa_I15016] which is B0032.ECFP
and
[http://partsregistry.org/wiki/index.php?title=Part:BBa_I15017 BBa_I15017 ]which is B0032.EYFP.
Tomorrow the green light promotor from PCR of 29_August should be inserted infront of the reporter proteins CFP and YFP.
blue light receptor
Miniprep
Investigators:Sophie
continue from: picking clones; 04.09.11
Testdigest showed no inserts
Ligation
Investigators: Sophie
again ligation as last time (02.08.11)
Lysis cassette
Troubleshooting of the modified Lysis genes K124017
Investigators:Theo
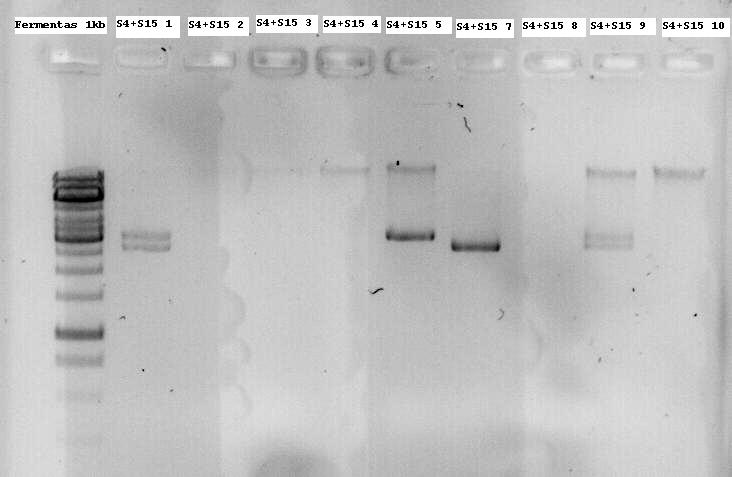
Minipreps of the first inoculations were applied on a 1% agarose gel without digesting them to see if they have an insert of the modified Lysis genes K124017. Nr1, 5 and 7 were sent for sequencing since the size seems to be about right.
- Note: S4 is the B0034 RBS (ribosome binding site), S15 is the non-modified K124017 Lysis Cassette without RBS
Precipitator
Ligation
| Name:Rüdiger | Date: 05.09. |
| Continue from Digestion Date 05.09 Name Rüdiger
Experiment | |
| Project Name: Precipitator | |
Procedure
PCR tube:
total volume 20 μl
- add H2O (17 μl -X-Y-Z)
- add 2 μl Ligase Buffer 10x
- add Insert 1, Insert 2(when proceeding from 3A digestion use 2 μl of each)
- add Vector (20ng needed. When proceeding from 3A digestion use 2 μl)
- Add 1 μl T4-DNA Ligase
- Incubate 10-30 min at room temperature
- heat for 20 minutes at 80°C
- store at -20°C or directly proceed to transformation
| Name of part | Ratio Insert:Vector
= 3:1 or 1:1 | Volume (μl) | |
| X insert 1 | |||
| Y insert 2 | |||
| Z vector | |||
| H2O |
Documentation:
Why are you doing this experiment? Where are your parts stored? Name the parts for ligation etc.
| All samples from last digestion.
|
Digestion
| Name:Rüdiger | Date: 05.09. |
| Continue from Digestion Date 31.08. Name Rüdiger
Experiment | |
| Project Name: Precipitator | |
Procedure
- add H2O (38μl-DNA )
- 5 μl NEB4 buffer (stored at iGEM’s, -20°C)
- 5 μl 10x BSA (used 1:10 diluted sample stored at iGEM’s, -20°C)
- DNA (500 ng) (Vector:Insert ratio 1:3 in following Ligation)
- 1 μl restriction enzymes (stored at iGEM’s, -20°C)
- heat for 1-2 hours 37°C (6 hours if time)
- heat for 20 minutes 80°C (inactivation of enzymes)
- keep at 4°C if you cannot continue
Measured DNA-concentration with Nanodrop to calculate the volume of DNA to do the digestion:
| Sample Name | DNA concentration (μg/μl) |
Restriction enzymes you need to cut the vector, insert1 and insert 2:
| Name | Vector (+Antarctic phosphatase 1 μl +AP buffer 5,6 μl) | Insert
(+Dpn1 1 μl) | Enzyme1 | Enzyme2 |
| 1 | PGEX David | 30 | EcoR1 | Bam H1 |
| 2 | PGEX David | 30 | EcoR1 | Bam H1 |
| 3 | PGEX David | 30 | EcoR1 | Bam H1 |
| 4 | C3 | 8 | EcoR1 | Spc1 |
| 5 | PET DUET | 12 | EcoR1 | Pst1 |
| 6 | PET DUET | 22
| EcoR1 | Pst1 |
| 7 | PET DUET | 20 | EcoR1 | Pst1 |
| 8 | C3 | 20 | EcoR1 | Spc1 |
| 9 | C3 | 20 | EcoR1 | Spc1 |
| 10 | C3 | 15 | EcoR1 | Spc1 |
Documentation:
Why are you doing this experiment? Where are the samples stored? Antibiotica resistance, vector used etc.
| Repeated digeston from 31.08.. |
Run a gel to verify if the part is cut out.
Miniprep
Investigators: Sophie
continue from experiment: Transformation; 03.09.11
testdigest showed no inserts
Trafo
Investigators. Sandra
Repeat of the trafo of:
- pGEX vector (Amp. resistance)
- pGEX vector (Amp. resistance)
- pGEX vector (Amp. resistance)
- pSB1C3
- pET Duet vector (Amp. resistance)
- pET Duet vector (Amp. resistance)
- pET Duet vector (Amp. resistance)
- pSB1C3
- pSB1C3
- pSB1C3
Trafo
Investigators. Sandra
Repeat of the trafo of:
 "
"
 Contact
Contact