Team:Freiburg/Notebook/11 August
From 2011.igem.org

Contents |
green light receptor
Skypephoned with the iGEM-Team Uppsala
Investigators:Theo, Rüdiger, Jakob
We talked to two guys (Lei Sun and Hamid Gavali) from the igem team uppsala. They gave us some informations and support, quite nice guys.
Informations:
- few Primer sequences
- Touchdown-pcr protocol
Thx a lot for that!!!
blue light receptor
PCR
| Name: Sophie
| Date: 11.8.11 |
| Continue from Experiment: new experiment
| |
| Project Name: changes in the LovTAP for 3A assembly | |
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl | H20 | Name |
| 10µl | 5x Phusion Buffer | of Primer |
| 2.5µl | Primer fw | |
| 2.5µl | Primer dw | |
| 1µl | dNTPs | of Template DNA |
| 1µl | DNA-Template | |
| 0.5 µl | Phusion (add in the end) |
What program do you use? Normal PCR with 60°C annealing temperature
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
How did you label the PCR-Product, where is it stored and what do you do next?
red light receptor
NAME OF YOUR EXPERIMENT
Investigators:NAME
Lysis cassette
NAME OF YOUR EXPERIMENT
Investigators:NAME
Precipitator
Tramsformation with inducible Promoter
Investigators: Sophie We didn't get any clones with both Plastic binding domain and Promoter-RBS inside, the Pbd seems to be toxic when expressed.So we want to try if it is possible to get cells with an inducible Promoter and Pbd. In this case we could cultivate the cells and induce expression of the Pbd only shortly before lysis. As our Light-inducible Promoters are not finished we use the IPTG inducible Promoter BBa_J04500 (Plate4, 12A)
PCR
Investigators: Rüdiger
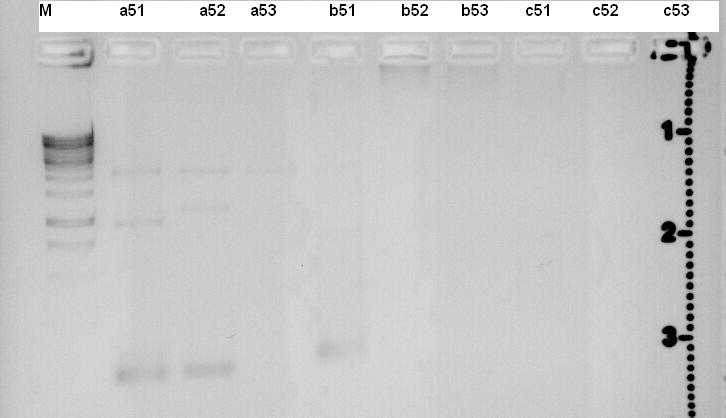
PCR of precipitator.
Primer fw:
- p51
- p52
- p53
Primer dw:
- p54
DNA template: S43
DMSO was used to reduce Tm about 5°C.
For the first a51, a52 and a53 6% DMSO was used. For b51, b52 and b53 9% Dmso was used. For c51, c52 and c53 12% Dmso was used.
Picking colonies
Investigators: Rüdiger
5 colonies were picked and incubated overnight in LB amp medium.
 "
"
 Contact
Contact