Team:UCL London/Research/Supercoiliology/Theory
From 2011.igem.org
The Theory of Gyrase
DNA gyrase was discovered in 1976 by a group of researchers (Martin Gellert et.al.) working at the National Institute of Health, Bethesda, Maryland, USA [1]. This prokaryotic enzyme is known to be the only one under its class (topoisomerase type II) and is the only topoisomerase capable of introducing negative supercoils into closed circular plasmid molecules. This unique negative supercoiling reaction requires magnesium ions and adenosine triphosphate (ATP) as the energy source. DNA gyrase therefore plays an important role in the growth as well as in the replication of E. coli.
A series of studies have reported that DNA gyrase relaxes the stress which is built up ahead of the gene transcription and DNA replication machinery. The stress of positive supercoils which accumulates downstream of these transcribing and replicating protein complexes is relieved by DNA gyrase molecules to allow for easier and obstruction-free propagation of the participating DNA or RNA polymerase complexes.
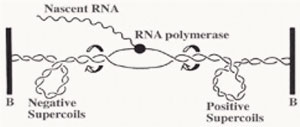
 Furthermore, negatively supercoiled plasmids also enables easier transition from the closed promoter complex to the open promoter complex and this accounts for higher transcriptional efficiency of the underwound B-DNA (right handed helix) molecule [2]. Although in theory, this indicates that all promoters will therefore have better transcriptional efficiency, but as usual there are specific exceptions.
Furthermore, negatively supercoiled plasmids also enables easier transition from the closed promoter complex to the open promoter complex and this accounts for higher transcriptional efficiency of the underwound B-DNA (right handed helix) molecule [2]. Although in theory, this indicates that all promoters will therefore have better transcriptional efficiency, but as usual there are specific exceptions.
In E.coli, the level of supercoiling actually acts as a regulatory mechanism for controlling the transcription and subsequent expression of gyrase molecules inside the cell. Gyrase genes are known to be repressed when the extent of negative supercoiling exceeds a certain threshold level. This molecular phenomenon is due to the spatial conformation adopted by the DNA sequences flanking the gyrase genes and promoters [2]. The transcription of gyrA and gyrB genes, which code for the subunits of DNA gyrase, is induced by relaxation of the DNA molecule. It has been proposed that promoter clearance instead of open complex formation is the actual rate-limiting step for these promoters.
In the presence of a high degree of negative supercoiling in the DNA molecule, the expression of DNA gyrase is seen to repressed while that of topoisomerase I seems to be upregulated automatically. This topoisomerase I is another enzyme which is responsible for the relaxation of negative supercoils in the DNA as opposed to the property of gyrase. The presence of this dual antagonistic enzymes therefore acts as a regulatory mechanism for maintaining a constant superhelical density (σ) and consistent topology within the E. coli chromosome. Under normal physiological conditions optimal to cell growth, the average superhelical density in an E coli cell is equivalent to 0.05 as indicated by the work carried out by Sinden et al. (1980) [3]. A model of this regulatory mechanism is displayed below. </html>
 "
"