Team:UCL London/Research/Supercoiliology/Model
From 2011.igem.org
Contents |
Modeling Gyrase Overexpression
The animation shows the functional E. coli DNA gyrase enzyme which consists of 2 GyrA subunits and 2 GyrB subunits. GyrB is the ATP consuming subunits and is yellow in colour in the animation. The blue coloured domains are the GyrA subunits whereby DNA breakage and resealing occurs, while the cyan colour domains are the C-Terminal domains of GyrA and are involved in the wrapping of DNA for allowing the correct progression of a full catalytic cycle in order to introduce negative supercoiling into the plasmid DNA molecule.
We developed a mathematical model to demonstrate the production of gyrase in a typical Top10 E.coli cell over the course of the cell´s doubling time. Additionally, we modeled the change in the expression of gyrase caused by the insertion of the gyrase expression cassette we designed and how that influenced the level of supercoiled plasmids in the cell over time.In order to model the expression of gyrase and the way it´s inhibited by the level of supercoiled plasmid in the cell, we used the equations found from P. R. Jensen et al. (1999)[5] to demonstrate the feedback mechanism of supercoiled plasmid by DNA gyrase and Topoisomerase 1 and the data on typical supercoiled DNA linking numbers from Barth, Dederich and Dedon (2009)[6].
For a regular Top10 E.coli cell
Assumptions:
1.1. There are 300 plasmids in a typical Top10 E.coli cell that have a length of 4000 nucleotide base pairs each on average, and 50 of which code for DNA gyrase.
1.2. All the plasmids that need to be expressed in the parent cell are present at t=0 of the cell´s life.
1.3. The plasmids coding for gyrase produce as much gyrase as the number of copies there are in within the cell for that particular plasmid.
1.4. All plasmids within this Top10 cell have the same copy number.
1.5. Both the subunits for gyrase A and gyrase B are expressed each plasmid coding for gyrase
E.coli cells have a doubling time of 40 minutes.
We made use of the transcription and translation rates of the plasmid coding for gyrase in order to ascertain the time taken for gyrase production in the cell.
1.6 Assume that transcription and translation of the gyrase encoding plasmids occur simultaneously and there are no limiting factors in the cell for these processes. That is to say that the ribosomes, transfer RNA (tRNA) and any enzymes required in the RNA forming complex are not limiting, allowing optimal, constant, translational and transcriptional rates.
1.7 Assume that within the cell the optimal conditions for growth are being met and maintained within the cell throughout its life cycle.
1.8 The overexpression of gyrase enzyme won´t slow down E.coli cell growth resulting in cell death.
For E.coli cells the rates of transcription and translation are 50 nucleotides/ second and 20 amino acids/ second (which corresponds to 60 nucleotides/ second) respectively. Since both of these reactions occur simultaneously, we must acknowledge that transcription is the rate determining step.
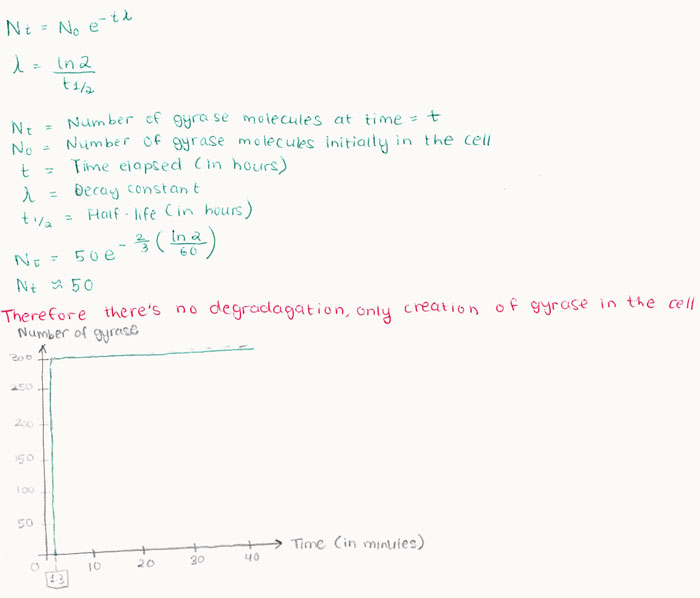
The first molecule of gyrase would be produced after 1.3 minutes. Assuming that all the plasmids coding for gyrase are fully expressed, at the end of the cell life (at t=40) there are meant to be 50 gyrase molecules present in the cell. Assuming no limiting factors during protein production, all 50 molecules of gyrase should be expressed simultaneously at t= 1.3 minutes. The rate of gyrase degradation also has to be accounted for in order to ascertain the total amount of gyrase that will be present in the cell. According to Reece and Maxwell (1991), gyrase has a half-life of 60 hours at 23 degrees Celsius at a dissociation constant of 0.1 to 0.5 nM. Due to the lack of experimental data which indicate the half life of the gyrase in vivo, we will assume that these are the conditions faced by the Top10 E.coli cell being modelled.
Using the following equations derived for determining half life we were able to ascertain the amount of gyrase molecules left in the cell by the time the parent cell was ready to divide into two daughter cells. It was found that amongst the total number of gyrase molecules in the cell, not a single one is fully degraded within the timespan of 40 minutes. This is to say that there is a steady increase in the amount of gyrase over time due to the transcription and translation of the gyrase encoding plasmids, until the cell has expressed the maximum amount of gyrase after which gyrase production levels off. Evidence of this is demonstrated graphically below:
Knowing the maximum amount of gyrase present in the cell is beneficial as it allows us to determine the exact time taken to produce adequate levels of supercoiling within the cell.
Supercoiling in an unmodified Top10 E.coli cell
For DNA molecules present within the cell, it is commonplace that one strand is always wound around the other forming the DNA double helix. In order for a DNA molecule to be supercoiled, it must form a closed loop with its ends fixed. Once its ends are covalently fixed together, it is in its relaxed topological state. This state can be described by the following equations[7]:
Lk = Tw+Wr
Lk=Linking number (Number of double helical turns in the original linear molecule)
Tw=The helical twist (Represents the way in which strands of DNA coil around each other about the axis of the DNA helix
Wr= superhelical turns (Measure of the contortion of the helix axis in space)
In a relaxed DNA plasmid, Lk is known as Lk(0) and is equal to Tw while Wr=0. Tw= N/h, h is the number of base pairs per turn of the helix and N is the number of base pairs in the DNA.
So for our plasmids Lk(0)= 381, and Wr=0.
The specific linking difference (σ) is number of turns added or removed relative to the total number of turns in the relaxed plasmid and thus showing the level of supercoiling.
σ = (Lk-Lk(0))/Lk(0)
The typical specific linking difference for prokaryotic cells is σ=-0.06 [7]so the linking number for the E.coli Top10 plasmids is 358.14. The turnover number for gyrase B is 1 coil/ gyrase/ second, that is a linking number of differences of ±2 per gyrase (which requires 2 molecules of ATP)/second.
The supercoiling of plasmid DNA (pDNA) to the standard amount within a prokaryotic cell requires a linking number deficit of 22.86 ≈ 23 per pDNA.
It takes 1.15 minutes in order to supercoil the entire plasmid content of the parent E.coli cell.
For a Top10 E.coli cell with the E.coili plasmid
Assumptions
2.1 The E.coili plasmid is a high copy number plasmid (ca 300 plasmids) and has a length of 5000 nucleotide base pairs.
2.2 The only difference between a normal Top10 E.coli cell and the Top10 cell that overexpresses gyrase is that the latter has the E.coili plasmid.
2.3 The E.coili plasmid will only express DNA gyrase.
2.4 The plasmids coding for gyrase produce as much gyrase as the number of copies there are in within the cell for that particular plasmid. This is because that the IPTG promoter which has been ligated into the E.coili plasmid does not subject to the natural chromosomal (regulatory mechanism explained in Supercoiliology Theory section).
2.5 All plasmids within this Top10 cell, exluding the E.coili plasmid have the same copy number.
Assumptions 1.1, 1.2, 1.5, 1.6, 1.7 and 1.8 still hold true.
The transcriptional and translational rates within the E.coli cell, as well as the growth conditions within the cell are to be held constant.
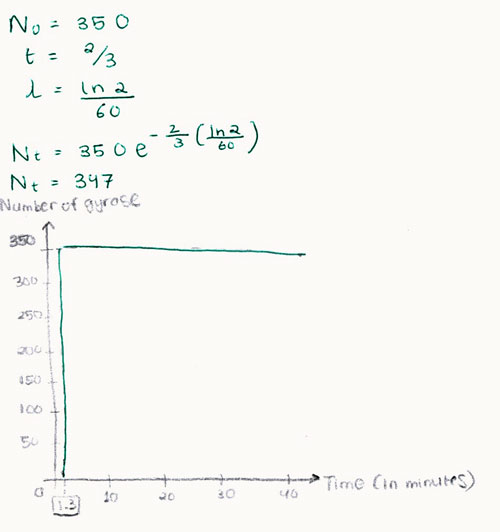
Therefore, the first gyrase molecule is produced after 1.3 minutes as well. Assuming no limiting factors during protein production, all 350 molecules of gyrase should be expressed simultaneously at t= 1.3 minutes. After 40 minutes, using the same calculations for half life as shown above, we get the gyrase degradation rate for this cell. It is found that after 40 minutes 347 out of a total of 350 gyrase molecules are surviving in the cell. Evidence of this is demonstrated graphically below:
Supercoiling in a modified Top10 cell with an E.coili plasmid
The typical specific linking difference for prokaryotic cells is σ=-0.06 so the linking number for the E.coli Top10 plasmids is 358.14. While the linking number for the high copy number plasmids encoding for gyrase is 447.44. The turnover number for gyrase B is 1 coil/ gyrase/ second, that is a linking number of differences of ±2 per gyrase (which requires 2 molecules of ATP)/second.
The supercoiling of plasmid DNA (pDNA) to the standard amount within a prokaryotic cell requires a linking number deficit of 28.66 ≈ 29 per E.coili plasmids and 22.86 ≈ 23 per pDNA.
It takes 0.37 minutes in order to supercoil the entire plasmid content of the parent E.coli cell. Even though there are a greater number of plasmids (which have more nucleotide base pairs), the increase in gyrase expressed in the cell allowed this larger amount of parental E.coli cell plasmid to be supercoiled more quickly than it was when the E.coili plasmid wasn´t being expressed. This demonstrates that the addition of our E.coili plasmid allows an overall increase in the speed of plasmid supercoiling to get a uniform amount of sc plasmid throughout the parental Top10 E.coli cell.
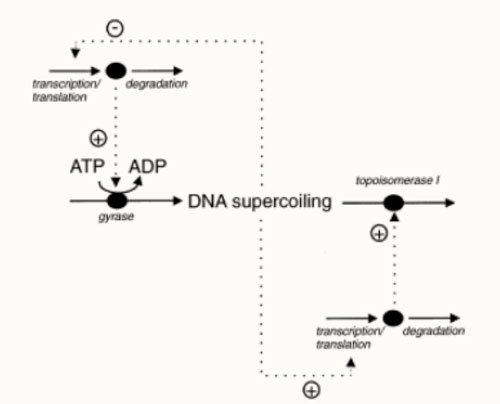
The following is a diagrammatic representation of the negative feedback mechanism for the production of gyrase within the cell according to P. R. Jensen et al. (1999). We were not able to create a mathematical model for this system since we lacked the kinetic data for DNA topoisomerase 1 as well as the fluctuating ATP/ ADP ratio according to set boundaries within the cell.
The upper level of this diagram represents the synthesis of gyrase, while the lower level represents the breakdown of gyrase and topoisomerase 1.
 "
"