Violacein
Courtesy of the Boeke Lab, our violacein biosynthesis genes VioA, VioB, VioC, VioD and VioE have been codon-optimized for yeast and designed to ligate with yeast specific promoters and UTR terminators. To assemble full expression cassettes for each gene, the genes have been fitted with flanking restriction sites:
Violacein Open Reading Frame (ORF) construct:
- XmaI site-BsaI site-Violacein ORF-inverted BsaI site-SphI site.
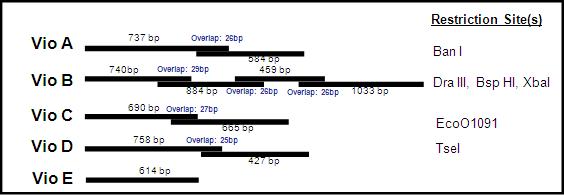
Depending on the above constructs size, we received it in pieces or 'building blocks'. VioA, C, D came in two building blocks. VioE was small enough to come in one piece, while VioB--around 3kb in length--came in 4 building blocks. Each building block overlaps its counterpart, and within the overlap region is a unique restriction enzyme site. By digesting both blocks with the right restriction enzyme, they can be ligated together into to make a full construct such as the one above.
The role of the flanking XmaI and SphI sites were added to easily insert the ORF into a vector double digested with XmaI and SphI. In addition to the Vio ORF's, the 12 promotors and UTRs have flanking BsaI sites as above. The flanking BsaI sites on the ORF are designed for addition of the promoter and the UTR in the right spot, thus creating a viable Violacein gene expression cassette designed for yeast.
BsaI operates differently from other restriction enzymes, which allows the design of a synthetic "sticky overhang of your choice. For further details, we have added a Request for Comment (RFC) dedicated to a form BsaI gene assembly called "Golden Gate Assembly."
 "
"