Team:Edinburgh/Experiments
From 2011.igem.org
(→malS) |
|||
| Line 11: | Line 11: | ||
==BioSandwich tests== | ==BioSandwich tests== | ||
| - | The BioSandwich system is outlined on [[Team:Edinburgh/BioSandwich|its own page]]. BioSandwich creates parts with homologous ends, but there are a number of ways in which the final assembly could be carried out. | + | The BioSandwich system is outlined on [[Team:Edinburgh/BioSandwich|its own page]]. BioSandwich creates parts with homologous ends, but there are a number of ways in which the final assembly could be carried out. We have tested a number of final assembly methods, by attempting to join a promoter and LacZ construct to a Yellow Fluorescent coding sequence. If successful, this will produce a polycistronic RNA transcript, coding for proteins that turn the bacteria blue (on Xgal) but also make it fluoresce yellow. |
| - | + | BioSandwich inserts short spacers between each part to be assembled. Note that after spacer2 we expect to see some extra bases before we reach the RFC10 suffix, because of the choice of vector. We can call these extra bases the "stuffing". So, the expected sequence is: | |
: '''Vector -- spacerT7 -- Plac,LacZ -- spacer1 -- EYFP -- spacer2 -- stuffing''' | : '''Vector -- spacerT7 -- Plac,LacZ -- spacer1 -- EYFP -- spacer2 -- stuffing''' | ||
| Line 20: | Line 20: | ||
From each plate we sequenced a colony with the correct phenotype (if there were any) and some colonies with incorrect phenotypes, in an attempt to understand what the failure modes were. | From each plate we sequenced a colony with the correct phenotype (if there were any) and some colonies with incorrect phenotypes, in an attempt to understand what the failure modes were. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===Ligation Independent Cloning=== | ||
| + | |||
| + | |||
| + | ===Overlap Extension PCR=== | ||
| + | |||
| + | This method produces many correct colonies, although it also produces many incorrect colonies. | ||
| + | |||
| + | Overlap Extension PCR can be performed using '''FIXME''' primers. The extension time should be calculated from the entire product size. The final product will not be circular, so can be run on a gel to test its length. Later, it will need to have its ends ligated. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
===BioSandwich with Gibson Assembly=== | ===BioSandwich with Gibson Assembly=== | ||
| - | * '''Colony gby [blue, yellow]''' - expected scar is missing at approx base 680 of the forward chromatogram. Also, the spacer ends with ATG and the part starts with ATG, but only one ATG is seen. In total 9 bases are missing. There is homology of "tatgg" at both ends of the deleted fragment. There is a point mutation at approx base 500 of the forward chromatogram. | + | Like, OEPCR, this method produces many correct colonies and many incorrect colonies. |
| - | * '''Colony gb [blue]''' - there is a deletion of about 80 bases at approx base 700 of the chromatogram. This has been caused by homology of a "gggcgagg" found at both ends of the deleted sequence. The 9 bases mentioned for '''gby''' are also missing. | + | |
| - | * '''Colony gwy [yellow]''' - seems correct except for a point mutation in the reverse chromatogram. The mutation is synonymous (ctg -> ttg). There's no clear reason why the colony wasn't blue. | + | The Gibson method was described by [http://www.nature.com/nmeth/journal/v6/n5/full/nmeth.1318.html Gibson ''et al'' (2009)]. A gentle introduction was written by the 2010 Cambridge iGEM team, and is found [https://2010.igem.org/Team:Cambridge/Gibson/Introduction here]. |
| + | |||
| + | We sequenced a colony with the correct phenotype, as well as others with incorrect phenotypes: | ||
| + | |||
| + | * '''Colony "gby" [blue, yellow]''' - expected scar is missing at approx base 680 of the forward chromatogram. Also, the spacer ends with ATG and the part starts with ATG, but only one ATG is seen. In total 9 bases are missing. There is homology of "tatgg" at both ends of the deleted fragment. There is a point mutation at approx base 500 of the forward chromatogram. | ||
| + | * '''Colony "gb" [blue]''' - there is a deletion of about 80 bases at approx base 700 of the chromatogram. This has been caused by homology of a "gggcgagg" found at both ends of the deleted sequence. The 9 bases mentioned for '''gby''' are also missing. | ||
| + | * '''Colony "gwy" [yellow]''' - seems correct except for a point mutation in the reverse chromatogram. The mutation is synonymous (ctg -> ttg). There's no clear reason why the colony wasn't blue. | ||
Lessons learned: bits of homology, even quite small, can cause problems with this method. | Lessons learned: bits of homology, even quite small, can cause problems with this method. | ||
| Line 33: | Line 61: | ||
[Different primers were used and spacer2 was not needed.] | [Different primers were used and spacer2 was not needed.] | ||
| - | * '''Colony bby [blue, yellow]''' - correct sequence. | + | * '''Colony "bby" [blue, yellow]''' - correct sequence. |
| - | * '''Colony bb [blue]''' - has a massive deletion (several hundred bases) in the EYFP gene after about 60 bases. There is partial homology between the start and end of the deletion: "tggtcgagctggac" vs "tggacgagctgtac". | + | * '''Colony "bb" [blue]''' - has a massive deletion (several hundred bases) in the EYFP gene after about 60 bases. There is partial homology between the start and end of the deletion: "tggtcgagctggac" vs "tggacgagctgtac". |
| - | * '''Colony bw [plain]''' - has spacerT7 joined to spacer1, and the same massive deletion as '''bb'''. | + | * '''Colony "bw" [plain]''' - has spacerT7 joined to spacer1, and the same massive deletion as '''bb'''. |
Again homology seems to be an issue. Parts joining randomly (in bw) is worrying. | Again homology seems to be an issue. Parts joining randomly (in bw) is worrying. | ||
| Line 41: | Line 69: | ||
===BioSandwich with Ligation Independent Cloning=== | ===BioSandwich with Ligation Independent Cloning=== | ||
| - | + | In our experience, this method has been only weakly successful. We have seen one correct colony in several attempts. | |
| + | |||
| + | To attempt the LIC method, simply mix the parts and vector (4 uL of each). Heat a beaker of water to 95 C, and float the reaction tube in it, and allow everything to cool. Afterwards, it may now be necessary to centrifuge the tube briefly to move the liquid back to the bottom. | ||
| - | * '''Colony b1 [blue, yellow]''' - sequence read is of low quality but things seem correct. | + | * '''Colony "b1" [blue, yellow]''' - sequence read is of low quality but things seem correct. |
| - | * '''Colony y2 [yellow]''' - appears to simply be <partinfo>BBa_K216011</partinfo> | + | * '''Colony "y2" [yellow]''' - appears to simply be <partinfo>BBa_K216011</partinfo>, the template for the initial PCR |
| - | * '''Colony n4 [plain]''' - sequence is unreadable. | + | * '''Colony "n4" [plain]''' - sequence is unreadable. |
The y2 colony is probably expressing a template for PCR which made it all the way through the various steps that came afterwards. | The y2 colony is probably expressing a template for PCR which made it all the way through the various steps that came afterwards. | ||
| Line 53: | Line 83: | ||
This is expected to fail as Ligation Independent Cloning requires long overhangs at the end of each part, not blunt ends. No correct colonies were seen. | This is expected to fail as Ligation Independent Cloning requires long overhangs at the end of each part, not blunt ends. No correct colonies were seen. | ||
| - | * '''Colony bb1 [blue]''' - seems to be <partinfo>BBa_J33207</partinfo> followed by part of ''E. coli'' ferrous iron transport gene A. | + | * '''Colony "bb1" [blue]''' - seems to be <partinfo>BBa_J33207</partinfo> followed by part of ''E. coli'' ferrous iron transport gene A. |
| - | * '''Colony bw2 [plain]''' - has spacerT7 followed by the stuffing. | + | * '''Colony "bw2" [plain]''' - has spacerT7 followed by the stuffing. |
| - | * '''Colony bw3 [plain]''' - has RFC10 prefix followed by RFC10 suffix, as if XbaI and SpeI sites have joined in a normal vector. | + | * '''Colony "bw3" [plain]''' - has RFC10 prefix followed by RFC10 suffix, as if XbaI and SpeI sites have joined in a normal vector. |
===BioSandwich with CPEC: Circular Polymerase Extension Cloning=== | ===BioSandwich with CPEC: Circular Polymerase Extension Cloning=== | ||
| - | * '''Colony cby [blue, yellow]''' - there is a single-base deletion near the start of the PlacLacZ part, with no obvious cause. It is too early to affect expression. The last few bases of spacer1 are missing, as well as the first few bases of the EYFP part (9 bases total). This is explained by homology of "tatgg". Otherwise seems correct. | + | * '''Colony "cby" [blue, yellow]''' - there is a single-base deletion near the start of the PlacLacZ part, with no obvious cause. It is too early to affect expression. The last few bases of spacer1 are missing, as well as the first few bases of the EYFP part (9 bases total). This is explained by homology of "tatgg". Otherwise seems correct. |
| - | * '''Colony cb [blue]''' - Normal until the end of spacer1, which is followed immediately by spacer2 and the stuffing. | + | * '''Colony "cb" [blue]''' - Normal until the end of spacer1, which is followed immediately by spacer2 and the stuffing. |
| - | * '''Colony cw [plain]''' - spacerT7 is followed immediately by a second copy of spacerT7, after which comes spacer2 and a second copy of spacer2. Then the stuffing. | + | * '''Colony "cw" [plain]''' - spacerT7 is followed immediately by a second copy of spacerT7, after which comes spacer2 and a second copy of spacer2. Then the stuffing. |
This gave some of the weirder results. | This gave some of the weirder results. | ||
Revision as of 17:53, 10 September 2011
Results
Actual results of experiments are placed here.
BioSandwich tests
The BioSandwich system is outlined on its own page. BioSandwich creates parts with homologous ends, but there are a number of ways in which the final assembly could be carried out. We have tested a number of final assembly methods, by attempting to join a promoter and LacZ construct to a Yellow Fluorescent coding sequence. If successful, this will produce a polycistronic RNA transcript, coding for proteins that turn the bacteria blue (on Xgal) but also make it fluoresce yellow.
BioSandwich inserts short spacers between each part to be assembled. Note that after spacer2 we expect to see some extra bases before we reach the RFC10 suffix, because of the choice of vector. We can call these extra bases the "stuffing". So, the expected sequence is:
- Vector -- spacerT7 -- Plac,LacZ -- spacer1 -- EYFP -- spacer2 -- stuffing
Successful assemblies are expected to generate blue colonies that fluoresce yellow under a blue light. We tried several assembly methods, all of which generated many colonies with the correct phenotype, but also many colonies with an incorrect phenotype...
From each plate we sequenced a colony with the correct phenotype (if there were any) and some colonies with incorrect phenotypes, in an attempt to understand what the failure modes were.
Ligation Independent Cloning
Overlap Extension PCR
This method produces many correct colonies, although it also produces many incorrect colonies.
Overlap Extension PCR can be performed using FIXME primers. The extension time should be calculated from the entire product size. The final product will not be circular, so can be run on a gel to test its length. Later, it will need to have its ends ligated.
BioSandwich with Gibson Assembly
Like, OEPCR, this method produces many correct colonies and many incorrect colonies.
The Gibson method was described by [http://www.nature.com/nmeth/journal/v6/n5/full/nmeth.1318.html Gibson et al (2009)]. A gentle introduction was written by the 2010 Cambridge iGEM team, and is found here.
We sequenced a colony with the correct phenotype, as well as others with incorrect phenotypes:
- Colony "gby" [blue, yellow] - expected scar is missing at approx base 680 of the forward chromatogram. Also, the spacer ends with ATG and the part starts with ATG, but only one ATG is seen. In total 9 bases are missing. There is homology of "tatgg" at both ends of the deleted fragment. There is a point mutation at approx base 500 of the forward chromatogram.
- Colony "gb" [blue] - there is a deletion of about 80 bases at approx base 700 of the chromatogram. This has been caused by homology of a "gggcgagg" found at both ends of the deleted sequence. The 9 bases mentioned for gby are also missing.
- Colony "gwy" [yellow] - seems correct except for a point mutation in the reverse chromatogram. The mutation is synonymous (ctg -> ttg). There's no clear reason why the colony wasn't blue.
Lessons learned: bits of homology, even quite small, can cause problems with this method.
BioSandwich with Overlap Extension PCR
[Different primers were used and spacer2 was not needed.]
- Colony "bby" [blue, yellow] - correct sequence.
- Colony "bb" [blue] - has a massive deletion (several hundred bases) in the EYFP gene after about 60 bases. There is partial homology between the start and end of the deletion: "tggtcgagctggac" vs "tggacgagctgtac".
- Colony "bw" [plain] - has spacerT7 joined to spacer1, and the same massive deletion as bb.
Again homology seems to be an issue. Parts joining randomly (in bw) is worrying.
BioSandwich with Ligation Independent Cloning
In our experience, this method has been only weakly successful. We have seen one correct colony in several attempts.
To attempt the LIC method, simply mix the parts and vector (4 uL of each). Heat a beaker of water to 95 C, and float the reaction tube in it, and allow everything to cool. Afterwards, it may now be necessary to centrifuge the tube briefly to move the liquid back to the bottom.
- Colony "b1" [blue, yellow] - sequence read is of low quality but things seem correct.
- Colony "y2" [yellow] - appears to simply be <partinfo>BBa_K216011</partinfo>, the template for the initial PCR
- Colony "n4" [plain] - sequence is unreadable.
The y2 colony is probably expressing a template for PCR which made it all the way through the various steps that came afterwards.
BioSandwich with Blunt-End Ligation Independent Cloning
This is expected to fail as Ligation Independent Cloning requires long overhangs at the end of each part, not blunt ends. No correct colonies were seen.
- Colony "bb1" [blue] - seems to be <partinfo>BBa_J33207</partinfo> followed by part of E. coli ferrous iron transport gene A.
- Colony "bw2" [plain] - has spacerT7 followed by the stuffing.
- Colony "bw3" [plain] - has RFC10 prefix followed by RFC10 suffix, as if XbaI and SpeI sites have joined in a normal vector.
BioSandwich with CPEC: Circular Polymerase Extension Cloning
- Colony "cby" [blue, yellow] - there is a single-base deletion near the start of the PlacLacZ part, with no obvious cause. It is too early to affect expression. The last few bases of spacer1 are missing, as well as the first few bases of the EYFP part (9 bases total). This is explained by homology of "tatgg". Otherwise seems correct.
- Colony "cb" [blue] - Normal until the end of spacer1, which is followed immediately by spacer2 and the stuffing.
- Colony "cw" [plain] - spacerT7 is followed immediately by a second copy of spacerT7, after which comes spacer2 and a second copy of spacer2. Then the stuffing.
This gave some of the weirder results.
malS
We placed malS (an E. coli amylase gene) under the control of the lac promoter to make part <partinfo>BBa_K523006</partinfo>.
- Photo here
INP-EYFP
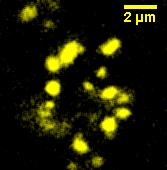
We made a fusion (<partinfo>BBa_K523013</partinfo>) of Ice Nucleation Protein and Enhanced Yellow Fluorescent Protein, and placed it under the control of the Lac promoter.
Under blue light, the cells fluoresced yellow, as shown on right. Unfortunately, our imaging technology was not capable of showing whether the fluorescence was localised to the outer membrane.
 "
"