Team:Edinburgh/Efficiency
From 2011.igem.org
Efficiency
We know from the C and Spatial Kappa models that, in principle, synergy can work. But, for the project to be truly feasible, display of cellulases via phage or INP must have benefits that outweigh the costs.
Consider this: for bacteria to produce phage or INP requires energy. This energy could have been spent producing extra copies of the cellulases. In order for the phage and cell display projects to make sense, the benefits of synergy must outweigh the cost of producing all these extra proteins.
This question can probably be investigated using simple maths and back-of-envelope calculations...
Contents |
Cell display
For the cell display system, the carrier protein INP (which is fused to a cellulase) is about two thirds the size of any of the cellulases. When fused to an INP protein, each cellulase thus has a final size about 66% above normal.
So if the synergystic system works at over 166% efficiency compared to the standard system where enzymes are free-floating in the media, cell display via INP makes sense.
Phage display
The situation with phage is more complex, and depends heavily on how many enzymes can be successfully displayed on a phage. Let us define some variables to help us think about the problem:
- S - how efficient the synergistic system is compared to the free-enzyme system e.g. 5 means 5 times as efficient.
- E - the energy cost to produce one enzyme
- N - the number of enzymes displayed on a phage
- P - the energy cost to produce one phage, not counting attached enzymes
The cost to the bacterium of producing a phage with all its displayed enzymes is thus E * N + P. In order for the system to be worth making, this cost must be less than the cost of producing enough free-floating enzymes to do the job equally well without synergy; this cost is S * E * N. Thus the system is worthwhile if:
- E * N + P < S * E * N
The critical value for N is therefore:
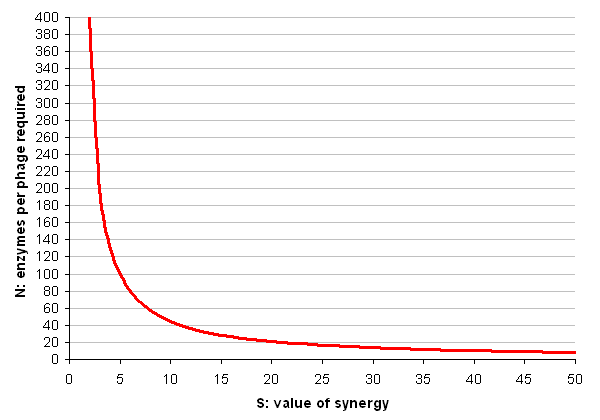
- N = P / ((S * E) - E)
Each cellulase is roughly 500 amino acids in size, but to produce one phage requires over 200,000 amino acids, as well as a significant amount of DNA synthesis (which we shall ignore). As for the value of synergy, a best-case scenario would be if the synergistic effects of having the enzymes in close proximity made them 50 times more efficient (this number is suggested and critiqued by Fontes and Gilbert, 2010).
So (finally the in silico part arrives) lets plug these numbers in and make a graph of N for all values of S between 2 and 50:
Conclusion
These results seem to suggest that the cell display system is the more feasible. Bearing in mind the dire warnings in the literature about how poorly large proteins display using conventional techniques, it seems the simple version of the phage system will probably not be useful in practice.
References
- Fontes CMGA, Gilbert HJ (2010) [http://www.annualreviews.org/doi/abs/10.1146/annurev-biochem-091208-085603 Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates]. Annual Review of Biochemistry 79: 655-81 (doi: 10.1146/annurev-biochem-091208-085603).
 "
"