Team:Edinburgh/Data
From 2011.igem.org
Revision as of 19:43, 2 September 2011 by Allancrossman (Talk | contribs)
Data Page
The iGEM rules require us to have simple illustrations of how our devices work and where the Parts function in the system; and links to the Registry for the parts/constructs for which we have produced data.
See the sample Data page.
Cell Surface Display System
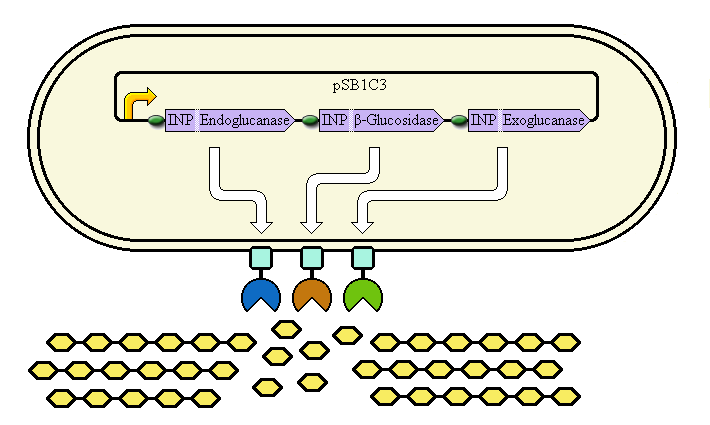
The completed system should contain:
A promoter (<partinfo>BBa_K523000</partinfo>) controlling:
an INP—Endoglucanase fusion (<partinfo>BBa_K523008</partinfo> + <partinfo>BBa_K523011</partinfo>)
an INP—β-glucosidase fusion (<partinfo>BBa_K523008</partinfo> + <partinfo>BBa_K523010</partinfo>)
an INP—Exoglucanase fusion (<partinfo>BBa_K523008</partinfo> + <partinfo>BBa_K523009</partinfo>)
Ribosome Binding Sites are indicated as green ovals.
Cellulose degradation is shown at top. In reality, tens of thousands of enzymes will cover the outer membrane in random places.
A test system to prove that <partinfo>BBa_K523008</partinfo> can be used to carry proteins to the outer membrane uses a fusion of INP to Yellow Fluorescent Protein (YFP) or the E. coli amylase MalS instead.
A promoter (<partinfo>BBa_K523000</partinfo>) controlling:
an INP—Endoglucanase fusion (<partinfo>BBa_K523008</partinfo> + <partinfo>BBa_K523011</partinfo>)
an INP—β-glucosidase fusion (<partinfo>BBa_K523008</partinfo> + <partinfo>BBa_K523010</partinfo>)
an INP—Exoglucanase fusion (<partinfo>BBa_K523008</partinfo> + <partinfo>BBa_K523009</partinfo>)
Ribosome Binding Sites are indicated as green ovals.
Cellulose degradation is shown at top. In reality, tens of thousands of enzymes will cover the outer membrane in random places.
A test system to prove that <partinfo>BBa_K523008</partinfo> can be used to carry proteins to the outer membrane uses a fusion of INP to Yellow Fluorescent Protein (YFP) or the E. coli amylase MalS instead.
Phage Display System
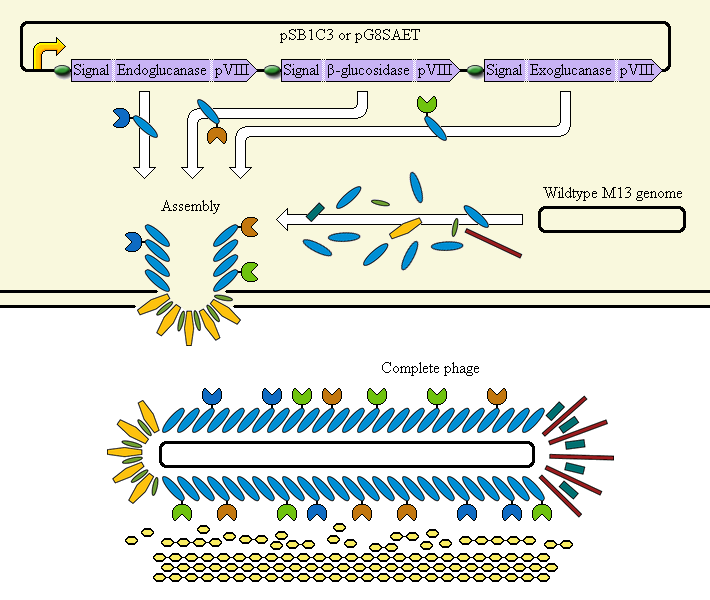
The completed system should contain:
A promoter (<partinfo>BBa_K523000</partinfo>) controlling:
an Endoglucanase—pVIII fusion
a β-glucosidase—pVIII fusion
an Exoglucanase—pVIII fusion
Ribosome Binding Sites are indicated as green ovals. "Signal" means a periplasmic signal sequence, directing the protein to the periplasm to be assembled into the phage.
A test system uses a fusion of pVIII to E. coli amylase MalS instead.
A promoter (<partinfo>BBa_K523000</partinfo>) controlling:
an Endoglucanase—pVIII fusion
a β-glucosidase—pVIII fusion
an Exoglucanase—pVIII fusion
Ribosome Binding Sites are indicated as green ovals. "Signal" means a periplasmic signal sequence, directing the protein to the periplasm to be assembled into the phage.
A test system uses a fusion of pVIII to E. coli amylase MalS instead.
 "
"