Team:Edinburgh/Data
From 2011.igem.org
(→Favourite BioBricks) |
(→Other BioBricks analysed) |
||
| Line 65: | Line 65: | ||
(More details of this are found at the dedicated [[Team:Edinburgh/Collaboration | Collaboration]] page. The information has also been added to the parts' "Experience" sections.) | (More details of this are found at the dedicated [[Team:Edinburgh/Collaboration | Collaboration]] page. The information has also been added to the parts' "Experience" sections.) | ||
| - | ; '''<partinfo>BBa_K415151</partinfo> | + | ; '''<partinfo>BBa_K415151</partinfo>: p8-GR1''' |
: Supposed to encode a fusion of the pVIII protein to a GR1 zipper. It actually encodes part of the ''E. coli'' aminopeptidase N gene. | : Supposed to encode a fusion of the pVIII protein to a GR1 zipper. It actually encodes part of the ''E. coli'' aminopeptidase N gene. | ||
| - | ; '''<partinfo>BBa_K392008</partinfo> | + | ; '''<partinfo>BBa_K392008</partinfo>: ''C. fimi'' beta-glucosidase''' |
: We discovered that this part almost certainly starts its coding sequence at the second ATG present, not the first. This is highly relevant information for anyone trying to tune its expression via use of custom Ribosome Binding Sites. | : We discovered that this part almost certainly starts its coding sequence at the second ATG present, not the first. This is highly relevant information for anyone trying to tune its expression via use of custom Ribosome Binding Sites. | ||
Revision as of 10:58, 8 September 2011
Data Overview
The iGEM rules require us to have simple illustrations of how our devices work and where the Parts function in the system; and links to the Registry for the parts/constructs for which we have produced data.
See the sample Data page.
Contents |
Overview
This page provides an overview of the purely biological aspects of our feasibility study.
Cell Surface Display System
(Further details are at the dedicated Cell Surface Display page.)
This system aims at achieving synergy between the enzymes by displaying them at high copy number on the cell's outer membrane. Ice Nucleation Protein is used as a carrier for display of the enzymes; it carries them to the outer membrane.
Schematic diagram
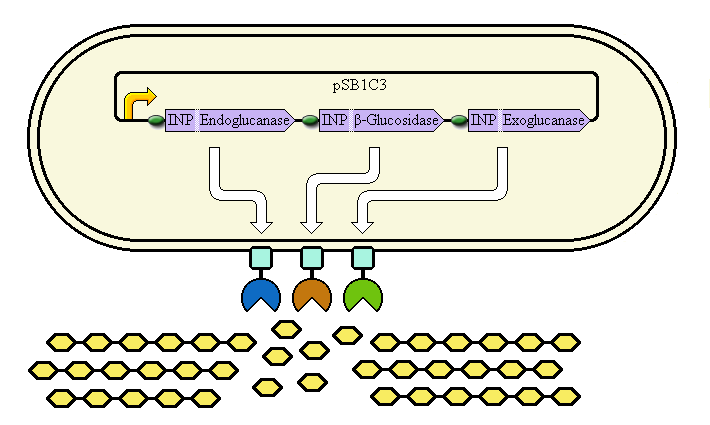
The completed system should contain:
A promoter (<partinfo>BBa_K523000</partinfo>) controlling:
an INP—Endoglucanase fusion (<partinfo>BBa_K523008</partinfo> + <partinfo>BBa_K523011</partinfo>)
an INP—β-glucosidase fusion (<partinfo>BBa_K523008</partinfo> + <partinfo>BBa_K523010</partinfo>)
an INP—Exoglucanase fusion (<partinfo>BBa_K523008</partinfo> + <partinfo>BBa_K523009</partinfo>)
Ribosome Binding Sites are indicated as green ovals.
Cellulose degradation is shown at the bottom. In reality, tens of thousands of enzymes will cover the outer membrane in random places.
A test system to prove that <partinfo>BBa_K523008</partinfo> can be used to carry proteins to the outer membrane uses a fusion of INP to Yellow Fluorescent Protein (YFP) or the E. coli amylase MalS (<partinfo>BBa_K523003</partinfo>) instead.
Phage Display System
(Further details are at the dedicated Phage Display page.)
This system aims at achieving synergy between the enzymes by displaying them on an M13 phage. The major coat protein pVIII is used as a carrier for display of the enzymes; it incorporates them into the phage.
Schematic diagram
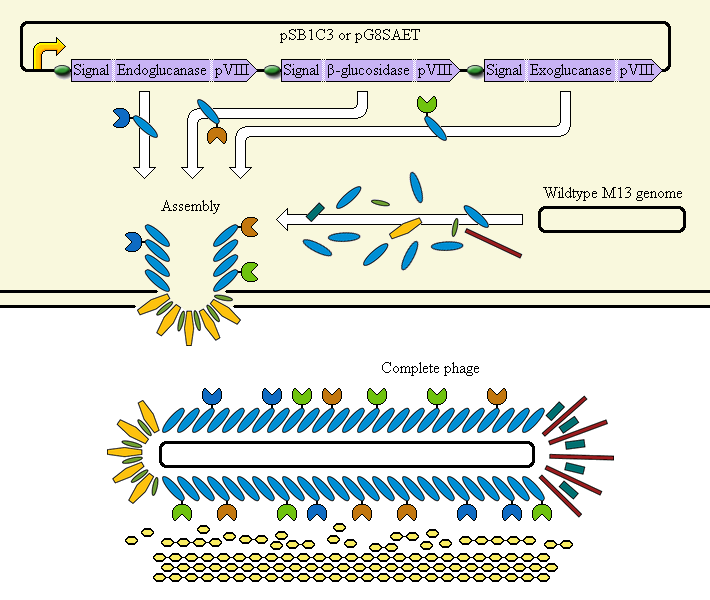
The completed system should contain:
A promoter (<partinfo>BBa_K523000</partinfo>) controlling:
an Endoglucanase—pVIII fusion
a β-glucosidase—pVIII fusion
an Exoglucanase—pVIII fusion
Ribosome Binding Sites are indicated as green ovals. "Signal" means a periplasmic signal sequence, directing the protein to the periplasm to be assembled into the phage.
A test system uses a fusion of pVIII to E. coli amylase MalS instead.
Favourite BioBricks
(A complete list of parts made during the project is found at the dedicated Parts page.)
- <partinfo>BBa_K523000</partinfo>: Plac + lacZ; with 5' BglII site
- Intended as a cloning vector (when present in pSB1C3), this part allows Blue/White selection of newly PCR'ed BioBricks. It encodes LacZα; new BioBricks that have successfully replaced the part will be white on plates containing IPTG and Xgal. Parts created in this way will also be compatible with the BioSandwich assembly protocol. We proved this part worked by using it to create several other BioBricks.
- <partinfo>BBa_K523013</partinfo>: Plac + INP-EYFP
- A fusion of Ice Nucleation Protein and Enhanced Yellow Fluorescent Protein under the control of the Lac promoter. Fluoresces yellow under blue light. May be localised to the outer membrane.
- <partinfo>BBa_K523014</partinfo>: Plac + bglX
- bglX is a cryptic E. coli β-glucosidase gene. We found this interesting and placed it under the control of the lac promoter.
Other BioBricks analysed
(More details of this are found at the dedicated Collaboration page. The information has also been added to the parts' "Experience" sections.)
- <partinfo>BBa_K415151</partinfo>: p8-GR1
- Supposed to encode a fusion of the pVIII protein to a GR1 zipper. It actually encodes part of the E. coli aminopeptidase N gene.
- <partinfo>BBa_K392008</partinfo>: C. fimi beta-glucosidase
- We discovered that this part almost certainly starts its coding sequence at the second ATG present, not the first. This is highly relevant information for anyone trying to tune its expression via use of custom Ribosome Binding Sites.
 "
"