Team:Bielefeld-Germany/SinceAmsterdam
From 2011.igem.org
JSchwarzhans (Talk | contribs) (→Selectivity) |
JSchwarzhans (Talk | contribs) (→Reductase?) |
||
| Line 16: | Line 16: | ||
The results of the experiment indicate that the bisdA | bisdB fusion protein has a '''high specificity for the degradation of BPA'''. In addition it is possible that the decrease in BPF and BPS concentration is due to internalization of those bisphenols or a endogenous enzyme of ''E. coli'' KRX and not the bisdA | bisdB fusion protein was responsible. It can be assumed that false positive signals because of BPF or BPS present in a sample are unlikely. | The results of the experiment indicate that the bisdA | bisdB fusion protein has a '''high specificity for the degradation of BPA'''. In addition it is possible that the decrease in BPF and BPS concentration is due to internalization of those bisphenols or a endogenous enzyme of ''E. coli'' KRX and not the bisdA | bisdB fusion protein was responsible. It can be assumed that false positive signals because of BPF or BPS present in a sample are unlikely. | ||
| - | === | + | ===BPA degradation with constructs containing FNR=== |
==S-layer== | ==S-layer== | ||
Revision as of 16:51, 28 October 2011

Contents |
BPA degradation
Specificity of bisphenol A degradation with E. coli
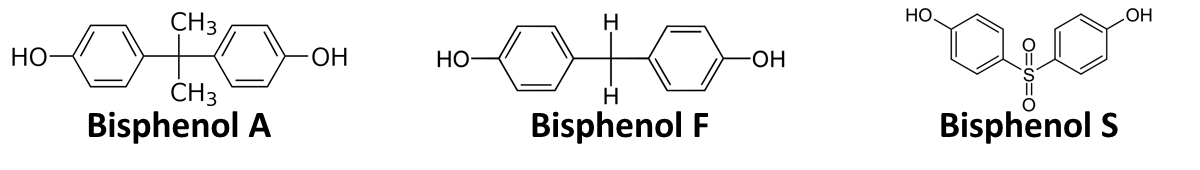
In order to access the specificity of the bisphenol A degradation by the bisdA | bisdB fusion protein we tested how well two similar bisphenols, bisphenol F (BPF) and bisphenol S (BPS), were employed. The structure of those bisphenols differs only in the chemical group linking the two phenols from that of bisphenol A (see Figure 7).
BPF and BPS are used in a broad range of applications that involve the use of polycarbonates or epoxy resins and thus can often be found were BPA is also present. Accordingly their presence is a potential disruptive factor that could lead to a false positive signal with our biosensor. This is especially true for BPS that in some cases is used as a substitute for BPA in baby bottles [http://www.nytimes.com/2011/04/18/business/global/18iht-rbog-plastic-18.html]. Studies concerning the environmental pollution with BPF ([http://www.sciencedirect.com/science/article/pii/S0043135401003670 Fromme et al. (2002)]) and the acute toxicity, mutagenicity and estrogenicity of BPF and BPS ([http://www.sciencedirect.com/science/article/pii/S0043135401003670 Chen et al. (2001)] and [http://toxsci.oxfordjournals.org/content/84/2/249 Kitamura et al. (2005)]) indicate their potential harmfulness but further research is needed to fully access their impact on human health.
E. coli KRX carrying genes for the bisdA | bisdB fusion protein behind the medium strong constitutive promoter <partinfo>J23110</partinfo> with RBS <partinfo>B0034</partinfo> was cultivated at 30 °C for 36 h with LB-Medium containing 120 mg L-1 BPA, BPF respectively BPS. The BPF and BPS concentration where determined with a HPLC using the same method as with BPA. Figure 8 shows the degradation of the respective bisphenol after 24 h of cultivation in percent.
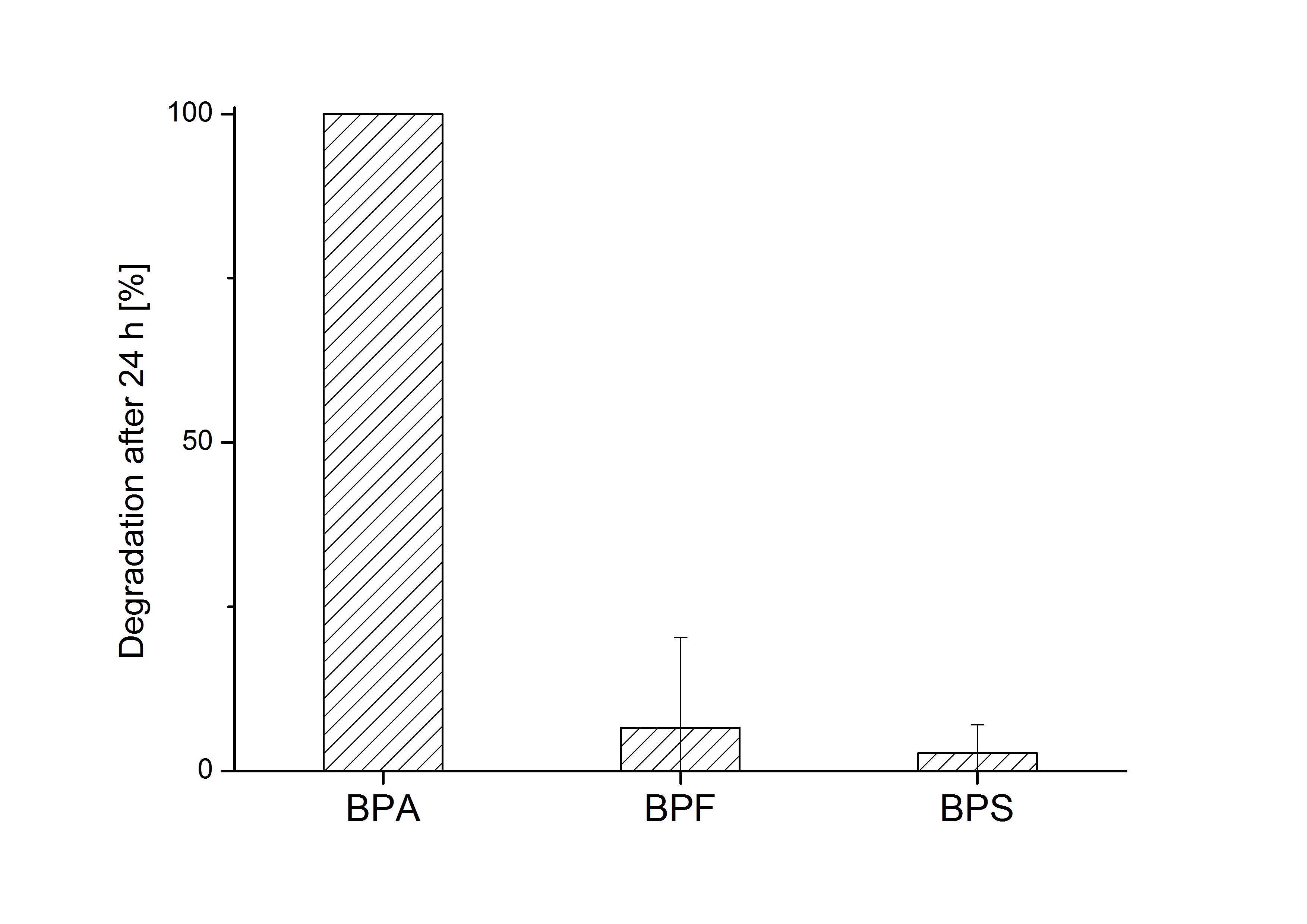
The results of the experiment indicate that the bisdA | bisdB fusion protein has a high specificity for the degradation of BPA. In addition it is possible that the decrease in BPF and BPS concentration is due to internalization of those bisphenols or a endogenous enzyme of E. coli KRX and not the bisdA | bisdB fusion protein was responsible. It can be assumed that false positive signals because of BPF or BPS present in a sample are unlikely.
 "
"