Team:Bielefeld-Germany/SinceAmsterdam
From 2011.igem.org
(→Selectivity) |
JSchwarzhans (Talk | contribs) (→Selectivity) |
||
| Line 2: | Line 2: | ||
==BPA degradation== | ==BPA degradation== | ||
| - | === | + | ===Specificity of bisphenol A degradation with ''E. coli''=== |
| + | |||
| + | In order to access the specificity of the bisphenol A degradation by the bisdA | bisdB fusion protein we tested how well two similar bisphenols, bisphenol F (BPF) and bisphenol S (BPS), were employed. The structure of those bisphenols differs only in the chemical group linking the two phenols from that of bisphenol A (see Figure 7). | ||
| + | |||
| + | [[Image:Bielefeld_2011_Bisphenols.png|700px|center|thumb| '''Figure 7: Chemical structure of BPA, BPF and BPS showing the different chemical groups linking the two phenols.''']] | ||
| + | |||
| + | BPF and BPS are used in a broad range of applications that involve the use of polycarbonates or epoxy resins and thus can often be found were BPA is also present. Accordingly their presence is a potential disruptive factor that could lead to a false positive signal with our biosensor. This is especially true for BPS that in some cases is used as a substitute for BPA in baby bottles [http://www.nytimes.com/2011/04/18/business/global/18iht-rbog-plastic-18.html]. Studies concerning the environmental pollution with BPF ([http://www.sciencedirect.com/science/article/pii/S0043135401003670 Fromme ''et al.'' (2002)]) and the acute toxicity, mutagenicity and estrogenicity of BPF and BPS ([http://www.sciencedirect.com/science/article/pii/S0043135401003670 Chen ''et al.'' (2001)] and [http://toxsci.oxfordjournals.org/content/84/2/249 Kitamura ''et al.'' (2005)]) indicate their potential harmfulness but further research is needed to fully access their impact on human health. | ||
| + | |||
| + | ''E. coli'' KRX carrying genes for the bisdA | bisdB fusion protein behind the medium strong constitutive promoter <partinfo>J23110</partinfo> with RBS <partinfo>B0034</partinfo> was cultivated at 30 °C for 36 h with LB-Medium containing 120 mg L<sup>-1</sup> BPA, BPF respectively BPS. The BPF and BPS concentration where determined with a HPLC using the same method as with BPA. Figure 8 shows the degradation of the respective bisphenol after 24 h of cultivation in percent. | ||
| + | |||
| + | [[Image:Bielefeld_2011_517_BPA-BPF-BPS_Degradation_24h_2.png|700px|center|thumb| '''Figure 8: Degradation of BPA, BPF and BPS after 24 h cultivation with ''E.coli'' KRX carrying genes for the bisdA <html>|</html> bisdB fusion protein behind the medium strong constitutive promoter <partinfo>J23110</partinfo> with RBS <partinfo>B0034</partinfo>. [[Team:Bielefeld-Germany/Protocols/Downstream-processing#Expression_of_bisphenol_A_degrading_BioBricks_in_E._coli | Cultivations]] were carried out at different temperatures in LB + Amp + bisphenol medium (starting concentration 120 mg L-1 BPA, BPF or BPS respectively) for 24 h in 300 mL shaking flasks without baffles with silicon plugs. Samples were taken at the end of the cultivation. Two biological replicates were analyzed. While BPA is fully degraded only a small fraction of BPF (~7%) and BPS (~3%) was degraded.''']] | ||
| + | |||
| + | The results of the experiment indicate that the bisdA | bisdB fusion protein has a '''high specificity for the degradation of BPA'''. In addition it is possible that the decrease in BPF and BPS concentration is due to internalization of those bisphenols or a endogenous enzyme of ''E. coli'' KRX and not the bisdA | bisdB fusion protein was responsible. It can be assumed that false positive signals because of BPF or BPS present in a sample are unlikely. | ||
| + | |||
===Reductase?=== | ===Reductase?=== | ||
Revision as of 16:35, 28 October 2011

Contents |
BPA degradation
Specificity of bisphenol A degradation with E. coli
In order to access the specificity of the bisphenol A degradation by the bisdA | bisdB fusion protein we tested how well two similar bisphenols, bisphenol F (BPF) and bisphenol S (BPS), were employed. The structure of those bisphenols differs only in the chemical group linking the two phenols from that of bisphenol A (see Figure 7).
BPF and BPS are used in a broad range of applications that involve the use of polycarbonates or epoxy resins and thus can often be found were BPA is also present. Accordingly their presence is a potential disruptive factor that could lead to a false positive signal with our biosensor. This is especially true for BPS that in some cases is used as a substitute for BPA in baby bottles [1]. Studies concerning the environmental pollution with BPF (Fromme et al. (2002)) and the acute toxicity, mutagenicity and estrogenicity of BPF and BPS (Chen et al. (2001) and Kitamura et al. (2005)) indicate their potential harmfulness but further research is needed to fully access their impact on human health.
E. coli KRX carrying genes for the bisdA | bisdB fusion protein behind the medium strong constitutive promoter <partinfo>J23110</partinfo> with RBS <partinfo>B0034</partinfo> was cultivated at 30 °C for 36 h with LB-Medium containing 120 mg L-1 BPA, BPF respectively BPS. The BPF and BPS concentration where determined with a HPLC using the same method as with BPA. Figure 8 shows the degradation of the respective bisphenol after 24 h of cultivation in percent.
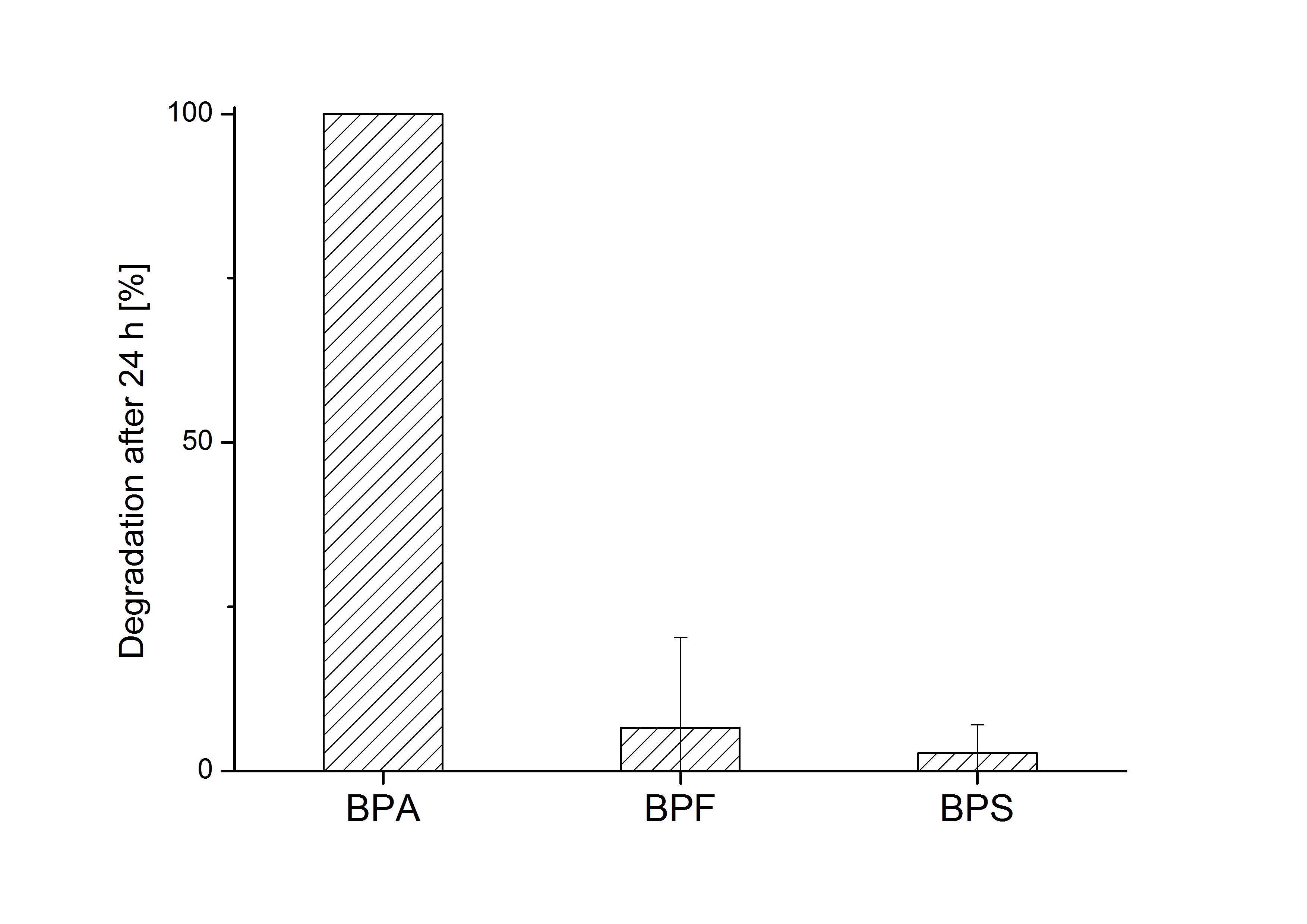
The results of the experiment indicate that the bisdA | bisdB fusion protein has a high specificity for the degradation of BPA. In addition it is possible that the decrease in BPF and BPS concentration is due to internalization of those bisphenols or a endogenous enzyme of E. coli KRX and not the bisdA | bisdB fusion protein was responsible. It can be assumed that false positive signals because of BPF or BPS present in a sample are unlikely.
 "
"