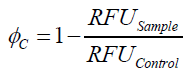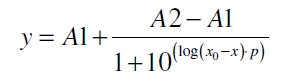Team:Bielefeld-Germany/Results/S-Layer/SgsE
From 2011.igem.org
(→Summary of advances) |
(→Summary of advances) |
||
| Line 3: | Line 3: | ||
=Summary of advances= | =Summary of advances= | ||
| - | - Primary experiments have been done to characterize the expression of [http://partsregistry.org/wiki/index.php/Part:BBa_K525305 K525305], which is a fusion of the sgsE gene ([http://partsregistry.org/wiki/index.php/Part:BBa_K525303 K525303]) with mCitrine ([http://partsregistry.org/Part:BBa_J18931 J18931]). Expression was observed through measurement of fluorescence intensity and optical density in induced and uninduced cultivations as well as in KRX. (See: [[Team:Bielefeld-Germany/Results/S-Layer/SgsE#Expression_in_E._coli|Expression in ''E. coli'']]). | + | - Primary experiments have been done to characterize the expression of [http://partsregistry.org/wiki/index.php/Part:BBa_K525305 K525305], which is a fusion of the sgsE gene ([http://partsregistry.org/wiki/index.php/Part:BBa_K525303 K525303]) with mCitrine ([http://partsregistry.org/Part:BBa_J18931 J18931]). Expression was observed through measurement of fluorescence intensity and optical density in induced and uninduced cultivations as well as in KRX. (See: [[Team:Bielefeld-Germany/Results/S-Layer/SgsE#Expression_in_E._coli|Expression in ''E. coli'']]). |
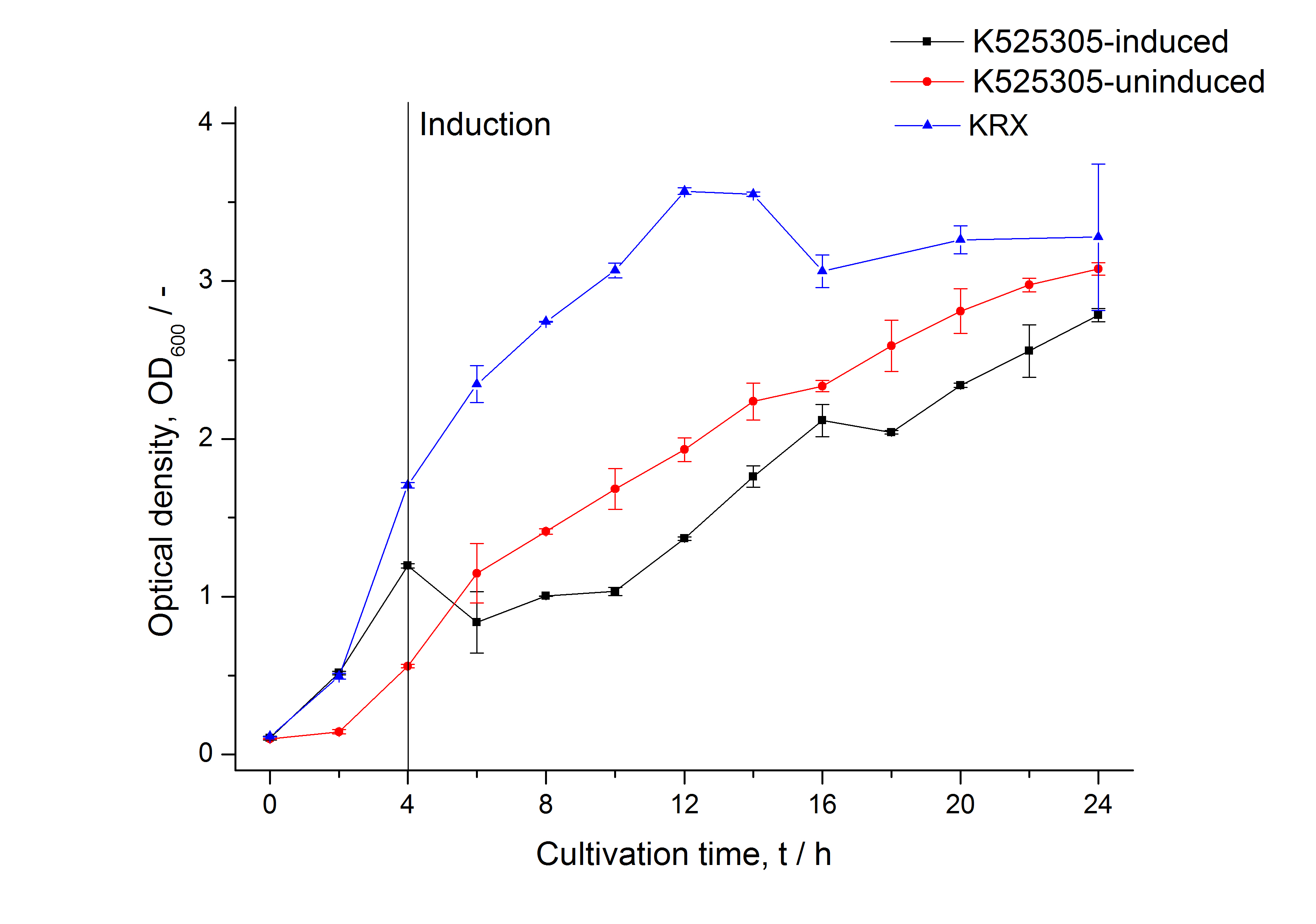
| - | - Knowing that the fusion protein forms inclusion bodies, next step was the development of a preferably easy purification. First purification steps after isolation and solubilization were different filtrations, using ultra- and diafiltration. The following dialyzation provided a solution of water-soluble fusion protein monomers which could further be used for recrystallization and coating experiments. (See: [[Team:Bielefeld-Germany/Results/S-Layer/SgsE#Purification_of_SgsE_fusion_protein|Purification of SgsE fusion protein inclusion bodies]]). | + | It could be shown that the fusion protein is translated in higher amount after induction. Although growth and increase in optical density are slightly higher in uninduced cultivations and in KRX without modifications, fluorescence increased clearly more in the induced cultivations. However fluorescence starts to decrease after 10 h of cultivation, indicating that optimal cultivation time is shorter than 12 h. |
| + | |||
| + | - Knowing that the fusion protein forms inclusion bodies, next step was the development of a preferably easy purification. First purification steps after isolation and solubilization were different filtrations, using ultra- and diafiltration. The following dialyzation provided a solution of water-soluble fusion protein monomers which could further be used for recrystallization and coating experiments. (See: [[Team:Bielefeld-Germany/Results/S-Layer/SgsE#Purification_of_SgsE_fusion_protein|Purification of SgsE fusion protein inclusion bodies]]). | ||
| + | |||
| + | It was found that during purification process a lot of protein is lost, especially after centrifugation following the cell lysis. Fluorescence is detergent fractions is lower as in the lysis fraction due to the denaturing nature of the used chemicals. After dialysis against HBSS some fluorescence could be regenerated. The methods of purification using different detergents led to the final purification strategy (See: [[Team:Bielefeld-Germany/Results/S-Layer/SgsE#Final_purification_strategy|final purification strategy]]. | ||
- After characterization of expression and development of an easy purification next goal was to coat beads. To characterize the immobilization behaviour fluorescence of the supernatant of the coating, the wash fraction of the beads as well as the beads themselves was measured. (See: [[Team:Bielefeld-Germany/Results/S-Layer/SgsE#Immobilization_behaviour|Immobilization behaviour]]). | - After characterization of expression and development of an easy purification next goal was to coat beads. To characterize the immobilization behaviour fluorescence of the supernatant of the coating, the wash fraction of the beads as well as the beads themselves was measured. (See: [[Team:Bielefeld-Germany/Results/S-Layer/SgsE#Immobilization_behaviour|Immobilization behaviour]]). | ||
<br/>Further immobilization kinetics were carried out with different bead concentration to find an optimal bead to protein ratio (See: [[Team:Bielefeld-Germany/Results/S-Layer/SgsE#Optimal_bead_to_protein_ratio_for_immobilization|bead to protein ratio]]). The data obtained from the experiments could also be fitted to a sigmoidal dose-response function, so a optimal bead to protein ratio could be determined. | <br/>Further immobilization kinetics were carried out with different bead concentration to find an optimal bead to protein ratio (See: [[Team:Bielefeld-Germany/Results/S-Layer/SgsE#Optimal_bead_to_protein_ratio_for_immobilization|bead to protein ratio]]). The data obtained from the experiments could also be fitted to a sigmoidal dose-response function, so a optimal bead to protein ratio could be determined. | ||
| + | |||
| + | The results of immobilization clearly indicate that a lot of protein did bind to the used silica beads, although some fluorescence could be measured in the supernatant of the coating fraction after immobilization and removing the coated beads. This is due to the high protein concentration used. The beads were satured with protein and still protein was left in the supernatant. Additionally the beads were washed several times with ddH<sub>2</sub>O to evaluate how firmly the immobilized protein is attached to the beads. Results are promising, showing only few fluorenscence in the wash fractions. | ||
| + | With immobilization kinetics using different bead concentrations an optimal bead to protein concentration could be calculated. A good silica bead conentration for immobilization of 100 µg protein is around 150 - 200 mg mL<sup>-1</sup>. | ||
=SgsE from ''Geobacillus stearothermophilus'' NRS 2004/3a= | =SgsE from ''Geobacillus stearothermophilus'' NRS 2004/3a= | ||
Revision as of 16:09, 27 October 2011


Contents |
Summary of advances
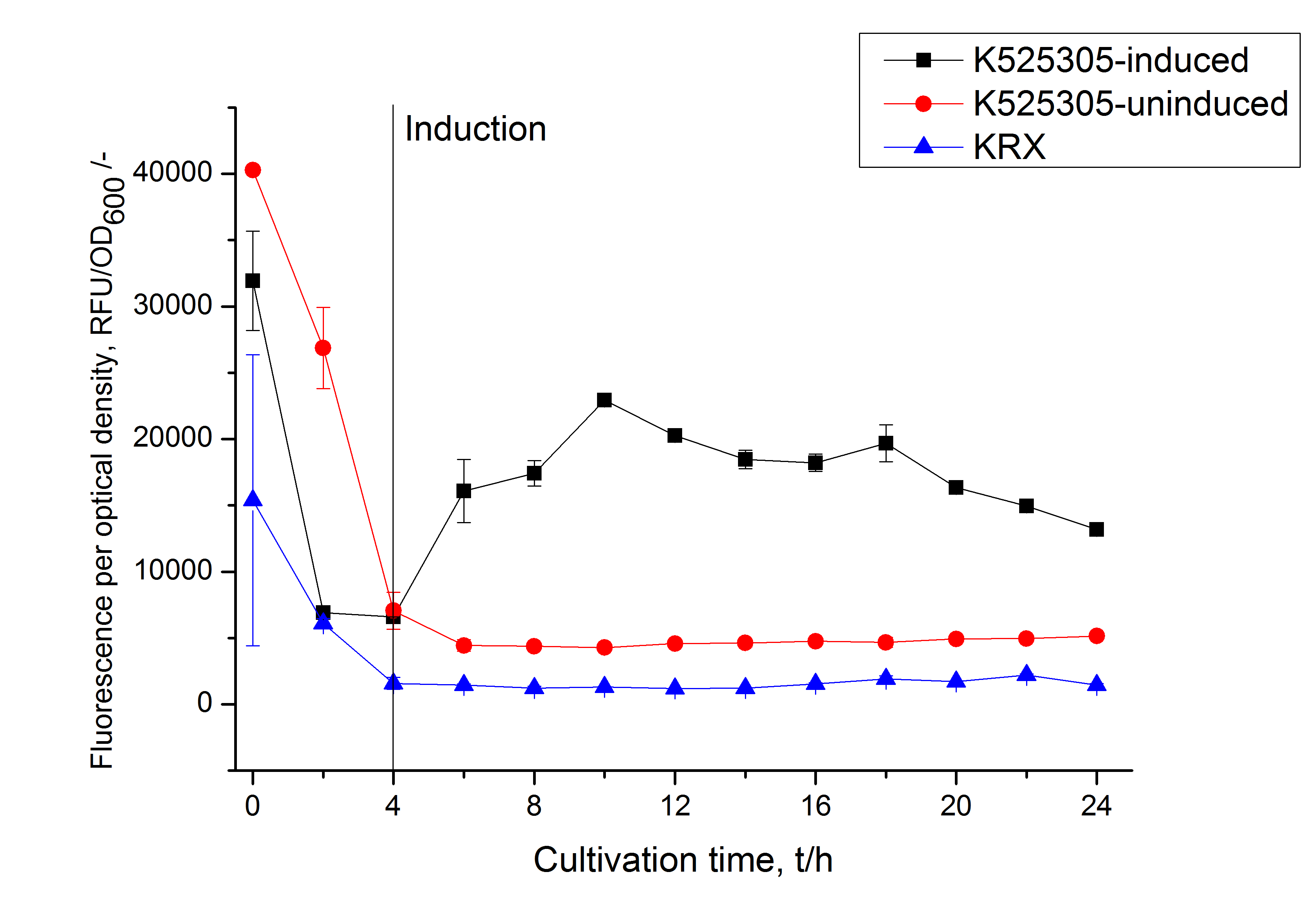
- Primary experiments have been done to characterize the expression of K525305, which is a fusion of the sgsE gene (K525303) with mCitrine (J18931). Expression was observed through measurement of fluorescence intensity and optical density in induced and uninduced cultivations as well as in KRX. (See: Expression in E. coli).
It could be shown that the fusion protein is translated in higher amount after induction. Although growth and increase in optical density are slightly higher in uninduced cultivations and in KRX without modifications, fluorescence increased clearly more in the induced cultivations. However fluorescence starts to decrease after 10 h of cultivation, indicating that optimal cultivation time is shorter than 12 h.
- Knowing that the fusion protein forms inclusion bodies, next step was the development of a preferably easy purification. First purification steps after isolation and solubilization were different filtrations, using ultra- and diafiltration. The following dialyzation provided a solution of water-soluble fusion protein monomers which could further be used for recrystallization and coating experiments. (See: Purification of SgsE fusion protein inclusion bodies).
It was found that during purification process a lot of protein is lost, especially after centrifugation following the cell lysis. Fluorescence is detergent fractions is lower as in the lysis fraction due to the denaturing nature of the used chemicals. After dialysis against HBSS some fluorescence could be regenerated. The methods of purification using different detergents led to the final purification strategy (See: final purification strategy.
- After characterization of expression and development of an easy purification next goal was to coat beads. To characterize the immobilization behaviour fluorescence of the supernatant of the coating, the wash fraction of the beads as well as the beads themselves was measured. (See: Immobilization behaviour).
Further immobilization kinetics were carried out with different bead concentration to find an optimal bead to protein ratio (See: bead to protein ratio). The data obtained from the experiments could also be fitted to a sigmoidal dose-response function, so a optimal bead to protein ratio could be determined.
The results of immobilization clearly indicate that a lot of protein did bind to the used silica beads, although some fluorescence could be measured in the supernatant of the coating fraction after immobilization and removing the coated beads. This is due to the high protein concentration used. The beads were satured with protein and still protein was left in the supernatant. Additionally the beads were washed several times with ddH2O to evaluate how firmly the immobilized protein is attached to the beads. Results are promising, showing only few fluorenscence in the wash fractions. With immobilization kinetics using different bead concentrations an optimal bead to protein concentration could be calculated. A good silica bead conentration for immobilization of 100 µg protein is around 150 - 200 mg mL-1.
SgsE from Geobacillus stearothermophilus NRS 2004/3a
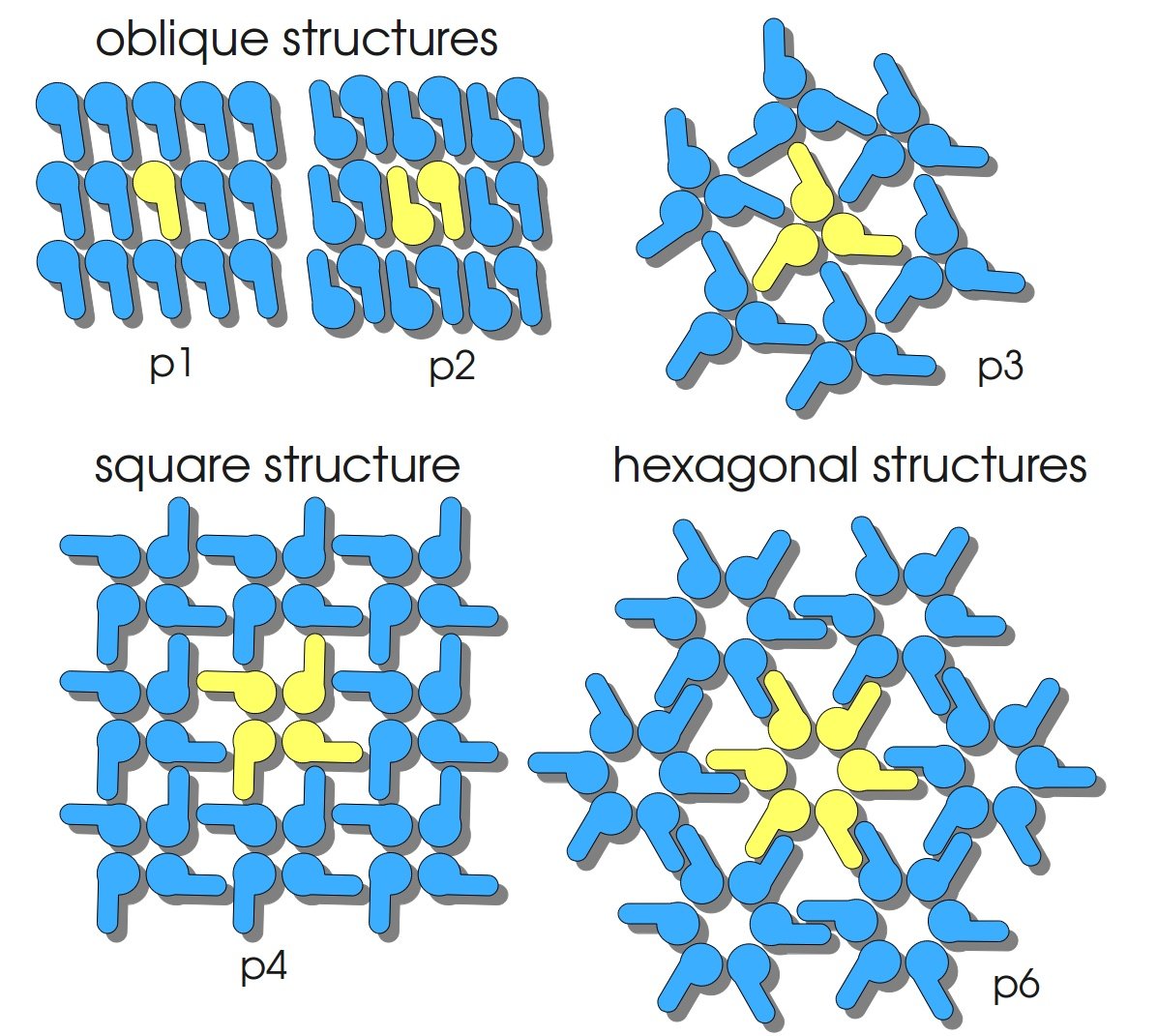
SgsE monomers are naturally assembled in a lattice with oblique symmetry (p2) (see fig. 2) exhibiting a well-defined periodicity and distances of 9.4 – 11.6 nm between the proteinaceous subunits. The S-layer protein SgsE of Geobacillus stearothermophilus NRS 2004/3a consists of 903 amino acids, including a 30 amino acid signal peptide (SLH-domain) at the amino-terminus. The carboxy-terminal part of SgsE is the larger part of the protein, encoding the self-assembly information. The protein is formed by the sgsE gene, has a calculated mass of 93.7 kDa and an isoelectric point of 6.1. When isolated SgsE maintains its ability to self-assemble, and depending on salt concentration, duration of dialysis to remove the detergent and its amino acid sequence it builds up five types of self-assembly products. These products are formed like flat sheets and cylinders (Schäffer et al., 2007).
Expression in E. coli
The SgsE gene under the control of a T7 / lac promoter (<partinfo>K525303</partinfo>) was fused to mCitrine (BBa_J18931) using Freiburg BioBrick assembly for characterization experiments.
The SgsE|mCitrine fusion protein was overexpressed in E. coli KRX after induction of T7 polymerase by supplementation of 0.1 % L-rhamnose and 1 mM IPTG using the autinduction protocol by Promega.
Purification of SgsE fusion protein
As observed in the analysis of the cultivations with expression of SgsE | mCitrine fusion proteins, these proteins form inclusion bodies in E. coli. Inclusion bodies have the advantage that they are relatively easy to clean-up and are resistant to proteases. So the first purification step is to isolate and solubilize the inclusion bodies. This step is followed by two filtrations (300 kDa UF and 100 kDa DF/UF) to further concentrate and purify the S-layer proteins. After the filtrations, the remaining protein solution is dialyzed against ddH2O for 18 h at 4 °C in the dark. The dialysis leads to a precipitation of the water-insoluble proteins. After centrifugation of the dialysate the water-soluble S-layer monomers remain in the supernatant and can be used for recrystallization experiments.
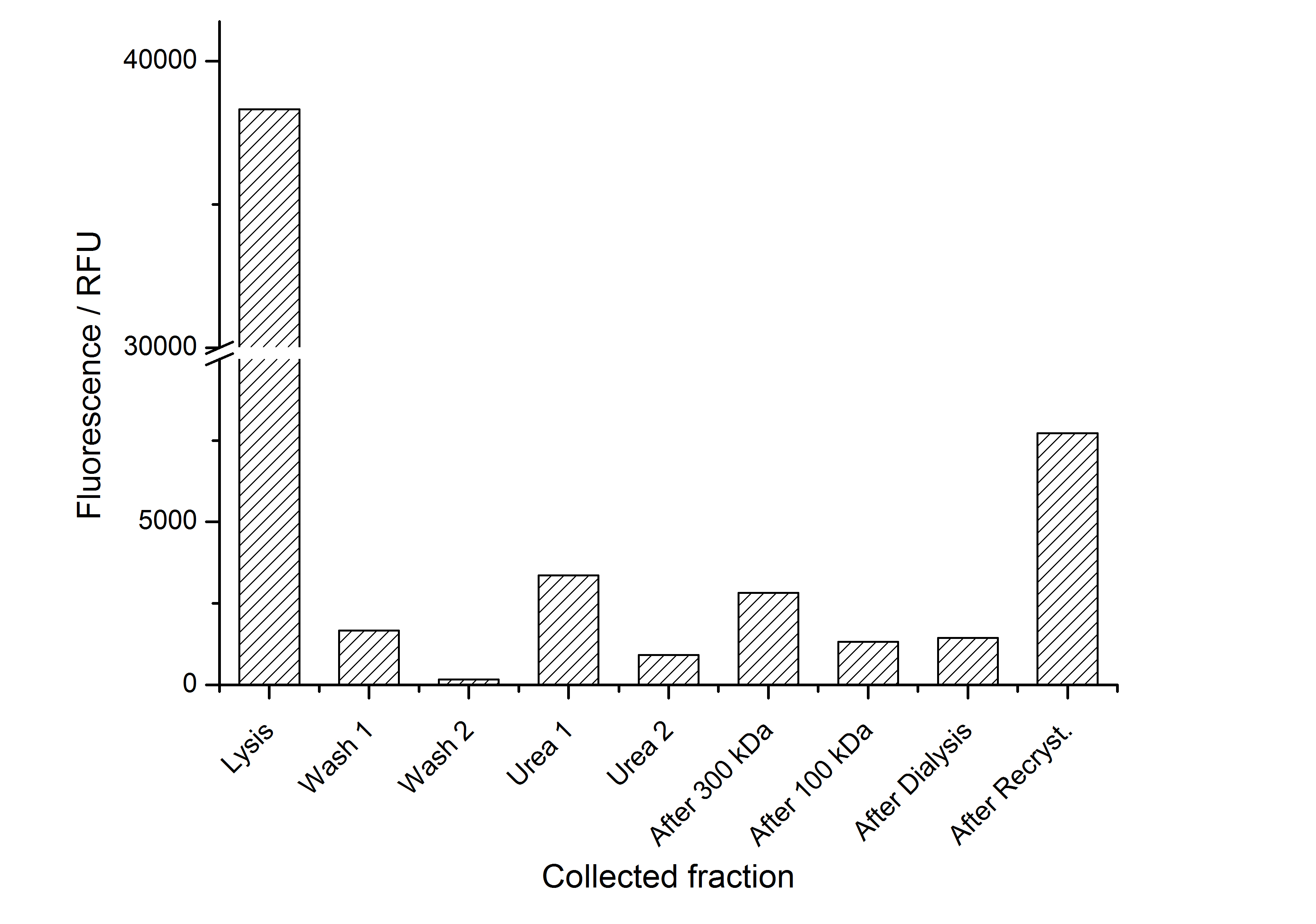
The fluorescence of the collected fractions of this purification strategy is shown in the following figure 3:
A lot of protein is lost during the purification especially after centrifugation steps. The fluorescence in the urea containing fractions is lowered due to denaturation of the fluorescent protein. Some fluorescence could be regenerated by the recrystallization in HBSS. This purification strategy is very simple and can be carried out by nearly everyone in any lab being one first step to enable real do it yourself nanobiotechnology.
Final purification strategy
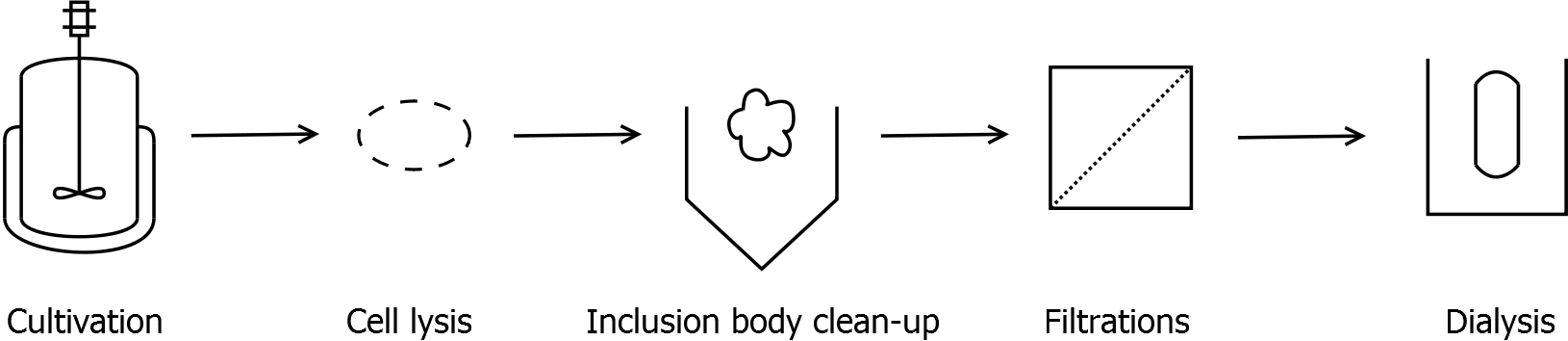
Scheme of purification strategy for SgsE (fusion) proteins:
First, SgsE is expressed in E. coli under the control of a T7 / lac promoter for separation of growth and production phase due to metabolic stress of the S-layer expression. Because the SgsE protein is forming inclusion bodies in E. coli, an inclusion body purification with urea follows the cell lysis. The S-layers are further concentrated and purified by two ultrafiltration / diafiltration steps (300 kDa and 100 kDa) and afterwards dialysed against water leading to the precipitation of water-insoluble proteins. The supernatant contains the monomeric SgsE solution.
Click for detailed information
Immobilization behaviour
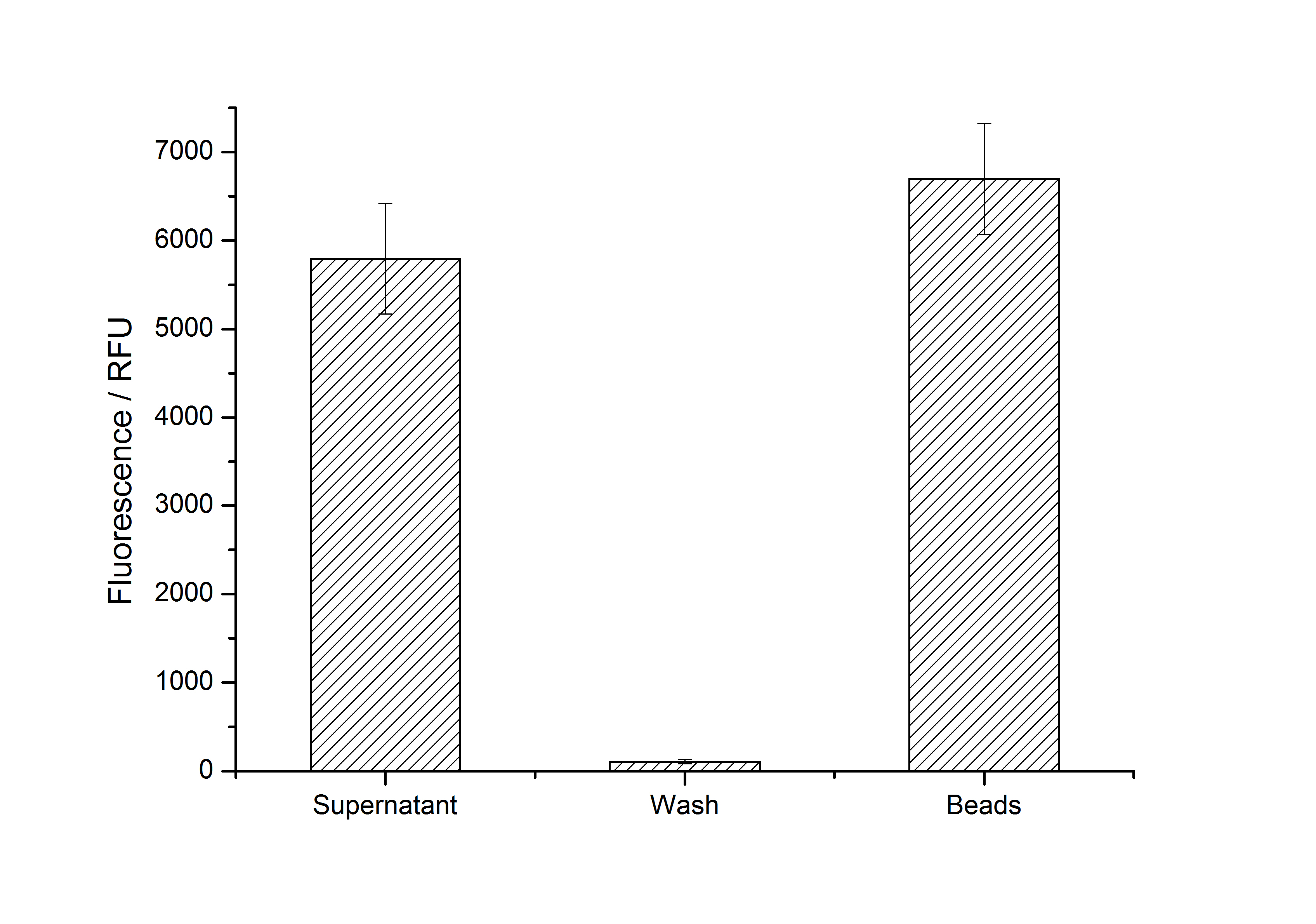
After purification, solutions of monomeric SgsE S-layer proteins can be recrystallized and immobilized on silicon dioxide beads in HBSS (Hank's buffered saline solution). After the recrystallization procedure the beads are washed with and stored in ddH2O at 4 °C in the dark. The fluorescence of the collected fractions of a recrystallization experiment with <partinfo>K525305</partinfo> are shown in fig. 4. 100 mg beads were coated with 100 µg of protein. The figure shows, that not all of the protein is immobilized on the beads (supernatant fraction) but the immobilization is pretty stable (very low fluorescence in the wash). After the immobilization, the beads show a high fluorescence indicating the binding of the SgsE | mCitrine fusion protein.
Optimal bead to protein ratio for immobilization
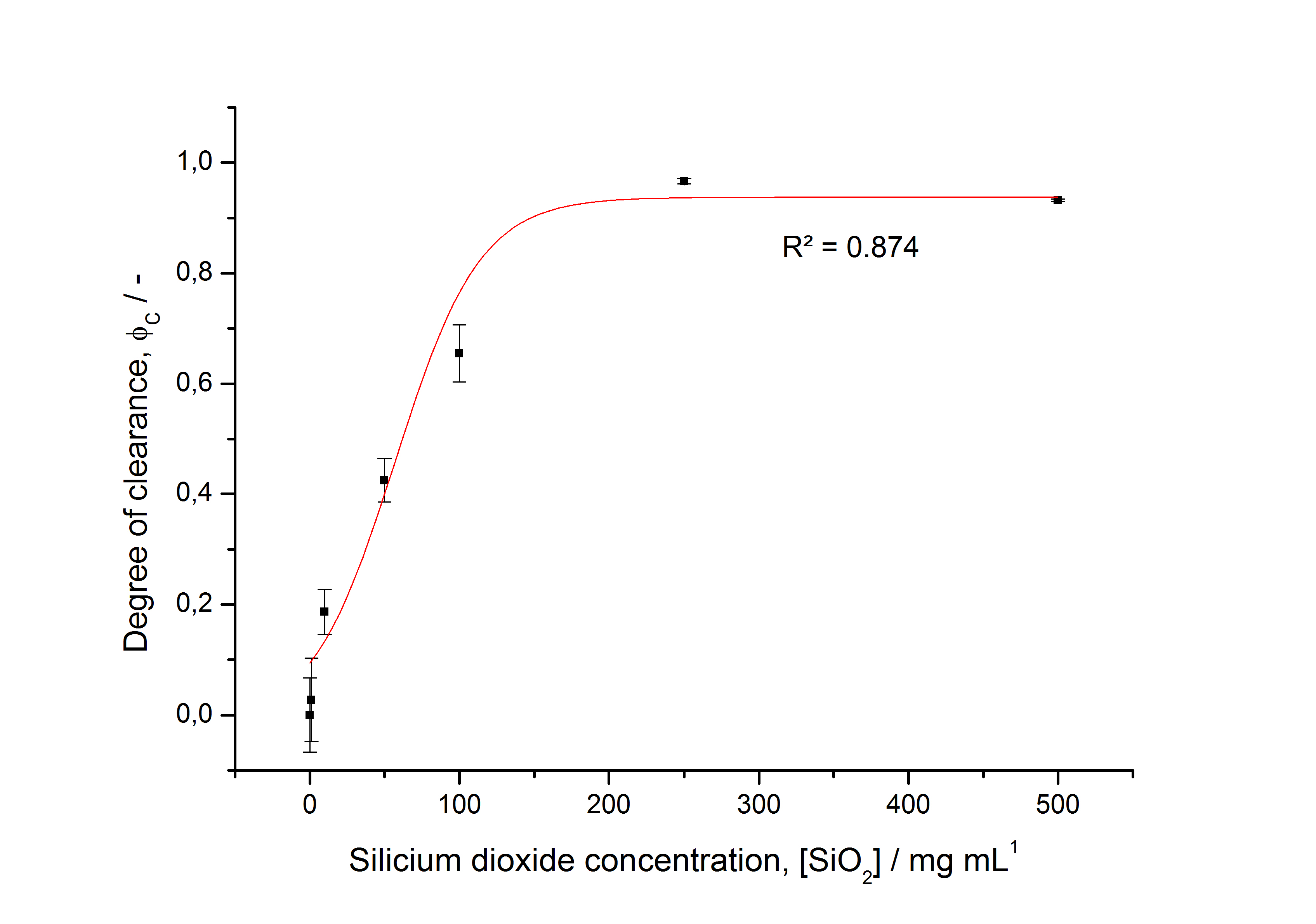
To determine the optimal ratio of silica beads to protein for immobilization, the degree of clearance ϕC in the supernatant is calculated and plotted against the concentration of silica beads used in the accordant immobilization experiment (compare fig. 5):
The data was collected in three independent experiments. The fluorescence of the samples was measured in the supernatant of the immobilization experiment after centrifugation of the silica beads. The fluorescence of the control was measured in a sample which was treated exactly like the others but no silica beads were added. 100 µg protein was used for one immobilization experiment. The data was fitted with a sigmoidal dose-response function of the form
with the Hill coefficient p, the bottom asymptote A1, the top asymptote A2 and the switch point log(x0) (R² = 0.874).
The fit indicates that a good silica concentration for 100 µg of protein is 150 - 200 mg mL-1. This set-up leads to saturated beads with low waste of protein. So a good protein / bead ratio to work with is 5 - 7 * 10-4.
 "
"