Team:Bielefeld-Germany/Results/S-Layer
From 2011.igem.org
(→The S-layer protein PS2 of Corynbacterium glutamicum) |
(→The S-layer protein PS2 of Corynbacterium glutamicum) |
||
| Line 50: | Line 50: | ||
|- | |- | ||
|style="border-style: solid; border-width: 0 1px 0 0"| [http://partsregistry.org/Part:BBa_K525121 K525121] | |style="border-style: solid; border-width: 0 1px 0 0"| [http://partsregistry.org/Part:BBa_K525121 K525121] | ||
| - | |style="border-style: solid; border-width: 0 1px 0 0"| [[Team:Bielefeld-Germany/Results/S-Layer/CspB_CG#Identification_and_localisation|CspB with TAT-sequence and lipid anchor]] | + | |style="border-style: solid; border-width: 0 1px 0 0"| [[Team:Bielefeld-Germany/Results/S-Layer/CspB_CG#Identification_and_localisation|<html> </html>CspB with TAT-sequence and lipid anchor]] |
|style="border-style: solid; border-width: 0 1px 0 0"| | |style="border-style: solid; border-width: 0 1px 0 0"| | ||
|style="border-style: solid; border-width: 0 1px 0 0"| | |style="border-style: solid; border-width: 0 1px 0 0"| | ||
| Line 59: | Line 59: | ||
|- | |- | ||
|style="border-style: solid; border-width: 0 1px 0 0"| [http://partsregistry.org/Part:BBa_K525123 K525123] | |style="border-style: solid; border-width: 0 1px 0 0"| [http://partsregistry.org/Part:BBa_K525123 K525123] | ||
| - | |style="border-style: solid; border-width: 0 1px 0 0"| [[Team:Bielefeld-Germany/Results/S-Layer/CspB_CG#CspB_without_TAT-sequence_and_with_lipid_anchor|CspB without TAT-sequence and with lipid anchor <html> </html>]] | + | |style="border-style: solid; border-width: 0 1px 0 0"| [[Team:Bielefeld-Germany/Results/S-Layer/CspB_CG#CspB_without_TAT-sequence_and_with_lipid_anchor|<html> </html>CspB without TAT-sequence and with lipid anchor <html> </html>]] |
|style="border-style: solid; border-width: 0 1px 0 0"| | |style="border-style: solid; border-width: 0 1px 0 0"| | ||
|style="border-style: solid; border-width: 0 1px 0 0"| | |style="border-style: solid; border-width: 0 1px 0 0"| | ||
Revision as of 08:17, 28 October 2011


Contents |
S-layer in general
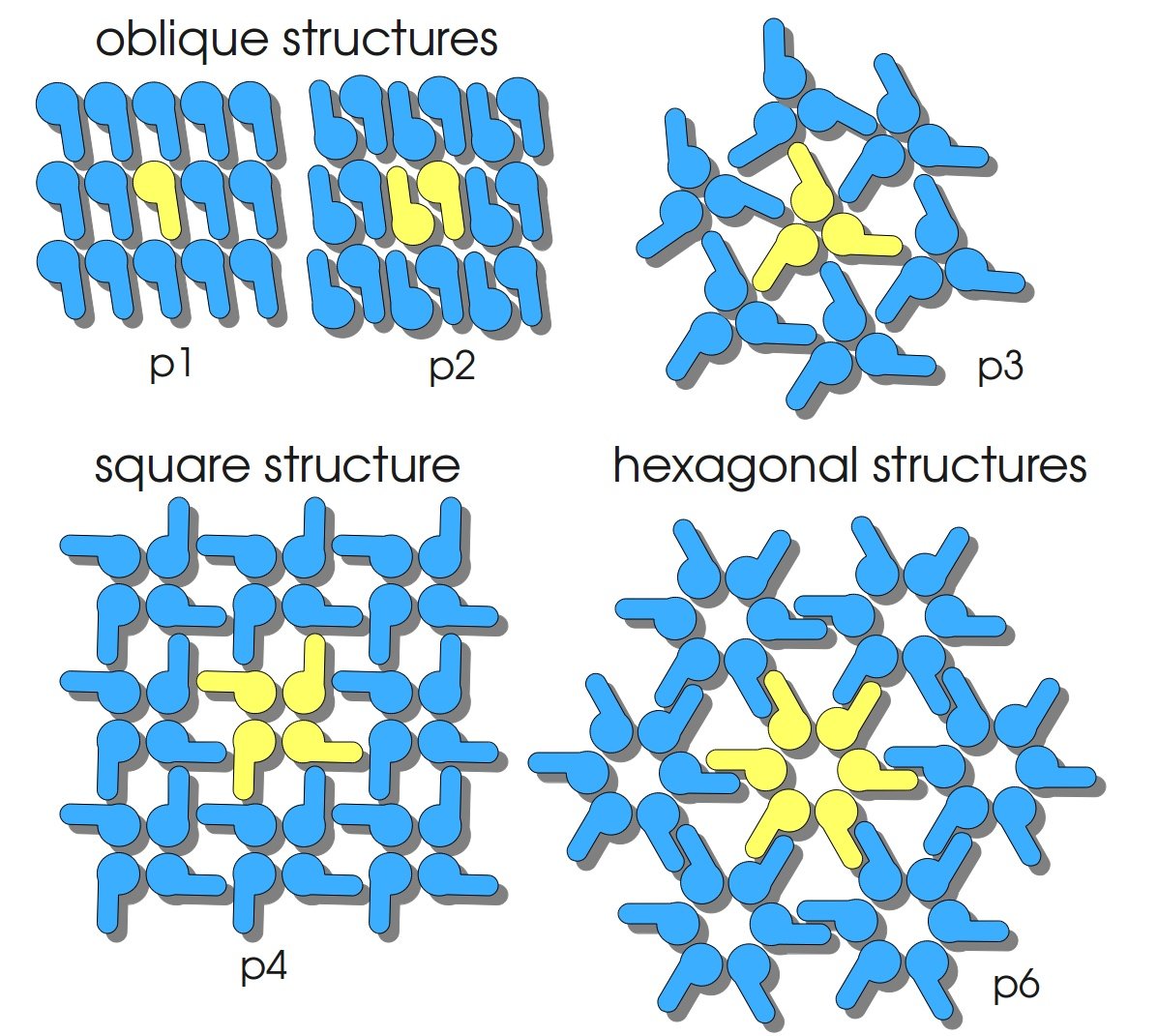
S-layer proteins have the ability to reassemble into two-dimensional crystals equal to the structures found on intact bacterial cells. Structures are periodic with very well defined distances between every protein unit. They posses porres of identical size and morphology and show equal physicochemical properties on each molecular unit. In solution, on solid supports and on phase-boundaries S-layer proteins can form self-assembly products or recrystallize into monomolecular layers. The protein subunits of S-layers are arranged in lattices with different symmetry: oblique (p1, p2), square (p4) or hexagonal (p3, p6) with a center-to-center spacing of the subunits of 3 – 35 nm (compare figure on the right). S-layers are highly porous with a porosity of 30 – 70 % (Sleytr et al., 2007).
The S-layer protein PS2 of Corynbacterium glutamicum
- Characterization of expression and induction of different variants of CspB fused with a monomeric RFP ((BBa_E1010). Expression was observed through measurement of optical density and fluorescence. Click on the construct to show the results.
| Biobrick Number | Construct | |
|---|---|---|
| K525121 | CspB with TAT-sequence and lipid anchor | |
| K525123 | CspB without TAT-sequence and with lipid anchor |
- Measurement of fluoresence to identify the location of the fusion protein in different fractions of the cells. Fractions were washed cells, the periplasm fraction after disruption of the periplasm, the supernatant of the medium after cultivation, the wash fraction of the pellet after lysis and wash with ddH2O, as well as the fractions after treating the pellet from lysis with different detergents.
- Identification of location of fusion protein in the cell through fractionation with different detergents and periplasmatic disruption. Identification of protein-containing fractions with fluorescence measurement and MALDI-TOF. Table below shows the supposed location of the fusion protein in the cell, depending on presence of lipid anchor and TAT-sequence. Click on the construct to show the results.
| Biobrick Number | Construct | Supernatant | Periplasm | Cell lysis | Integration in cell membrane | Inclusion bodies | |
|---|---|---|---|---|---|---|---|
| K525121 | CspB with TAT-sequence and lipid anchor | X | X | ||||
| K525123 | CspB without TAT-sequence and with lipid anchor | X | X |
The S-layer protein PS2 of Corynbacterium halotolerans
- Characterization of expression and induction of different variants of CspB from Corynebacterium halotolerans fused with a monomeric RFP ((BBa_E1010). Expression was observed through measurement of optical density and fluorescence. Click on the construct to show the results.
| Biobrick Number | Construct | |
|---|---|---|
| K525222 | CspB without TAT-sequence and lipid anchor | |
| K525223 | CspB without TAT-sequence and with lipid anchor | |
| K525224 | CspB with TAT-sequence and without lipid anchor |
- Measurement of fluoresence to identify the location of the fusion protein in different fractions of the cells. Fractions were washed cells, the periplasm fraction after disruption of the periplasm, the supernatant of the medium after cultivation, the wash fraction of the pellet after lysis and wash with ddH2O, as well as the fractions after treating the pellet from lysis with different detergents.
- Identification of location of fusion protein in the cell through fractionation with different detergents and periplasmatic disruption. Identification of protein-containing fractions with fluorescence measurement and MALDI-TOF. Table below shows the supposed location of the fusion protein in the cell, depending on presence of lipid anchor and TAT-sequence. Small x show small amounts of protein, large X show the main amount of protein. Click on the construct to show the results.
| Biobrick Number | Construct | Supernatant | Periplasm | Cell lysis | Integration in cell membrane | Inclusion bodies | |
|---|---|---|---|---|---|---|---|
| K525222 | CspB without TAT-sequence and lipid anchor | X | x | x | |||
| K525223 | CspB without TAT-sequence and with lipid anchor | x | x | ||||
| K525224 | CspB with TAT-sequence and without lipid anchor | X | x | x |
- Purification of the protein could be achieved through precipitation with ammonium sulfate, followed by ultra- and diafiltration. Salt concentration in the final anion exchange chromatography could be optimized to 400 mM NaCl (See: Purification and final strategy).
The S-layer protein SgsE of Geobacillus stearothermophilus NRS 2004/3a
The S-layer protein SbpA of Lysinibacillus sphaericus CCM 2177
Summary of results
Four different S-layer BioBricks with different lattice structures were created and sent to the partsregistry. The behaviour of these genes when expressed in E. coli were characterized and purification strategies for the expressed proteins were developed. Two purified fluorescent S-layer fusion proteins from different organisms were immobilized on beads, leading to a highly significant fluorescence enhancement of these beads (p < 10-14). Furthermore regarding the other two S-layers (CspB from Corynebacterium glutamicum and Corynebacterium halotolerans) we discovered that while expression with a lipid anchor resulted in an integration into the cellmembrane, the expression with a TAT-sequence resulted in a segregation into the medium. We also detected, that those S-layers seem to stabilize the biologically active conformation of mRFP. Furthermore we expressed and purified a fluorescent CspB fusion protein from C. halotolerans which has never been expressed in E. coli until now.
 "
"