Team:Bielefeld-Germany/Project/Background/BPA
From 2011.igem.org

Contents |
Bisphenol A
monomer, plastic, polycarbonate, epoxy resins, food containers, forbidden in EU and canada in baby flasks etc.
Bisphenol A and its effects on mammals
endocrine disruptor, estrogenic, teratogenic, infertility etc.
Bisphenol A degradation
There exist a lot of bacteria in nature that can degrade xenobiotic substances such as phenolic compounds or endocrine disruptors. In a lot of soil samples that were taken from contaminated soil to find organisms that do so, sphingomonads were extraordinarily often isolated (Stolz, 2009). In 2005, Sasaki et al. isolated a soil bacterium from the Sphingomonas genus which is able to degrade the endocrine disruptor bisphenol A (BPA) with a unique rate and efficiency compared to other BPA degrading organisms. This strain, called Sphingomonas bisphenolicum AO1, is able to completely decompose 120 mg BPA L-1 in about 6 hours while other strains need days of cultivation (e.g. Sphingomonas strain BP-7 isolated by Sakai et al. (2007)).
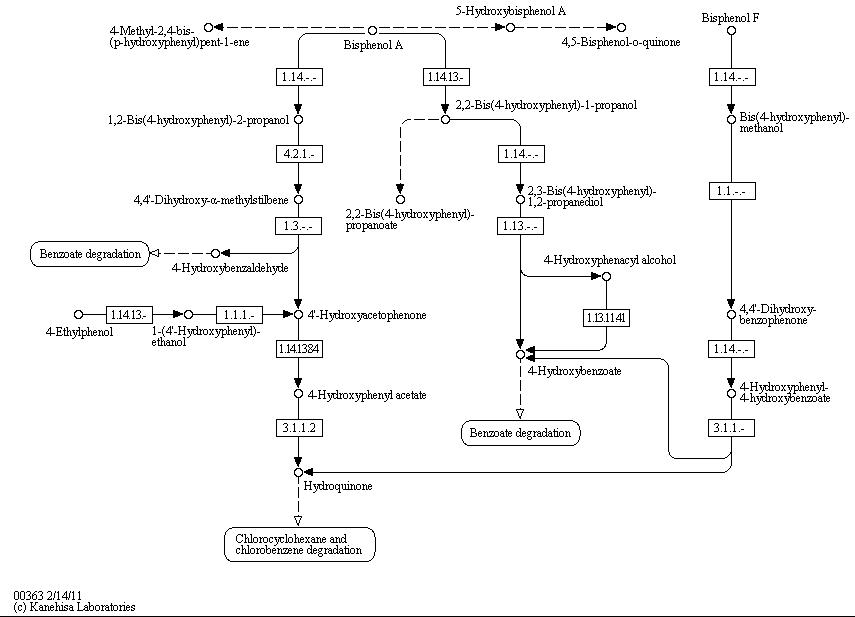
The full bisphenol A degradation pathway which is found in nature is shown in figure X (taken from KEGG database). BPA is metabolized to 4-Hydroxybenzaldehyde, 4'-Hydroxyacetophenone and 4-Hydroxybenzoate which can be used by some bacteria as carbon source. These and other metabolites of the BPA degradation pathway can be found in the supernatant of BPA containing cultivations of S. bisphenolicum AO1. They can grow on BPA as the only carbon source, too (Sasaki et al., 2005a).
Bisphenol A is mainly hydroxylated into the products 1,2-Bis(4-hydroxyphenyl)-2-propanol and 2,2-Bis(4-hydroxyphenyl)-1-propanol by some kind of oxidoreductase acting with NADH or NADPH. A total of three genes are responsible for this BPA hydroxylation: a cytochrome P450 (CYP, bisdB), a ferredoxin (Fd, bisdA) and a ferredoxin-NAD+ oxidoreductase (FNR) (Sasaki et al., 2005b). The three gene products act together to reduce BPA while oxidizing NADH + H+. The cytochrome P450 (BisdB) reduces the BPA and is oxidized during this reaction. BisdB in its oxidized status is reduced by the ferredoxin (BisdA) so it can reduce BPA again. The oxidized BisdA is reduced by a ferredoxin-NAD+ oxidoreductase consuming NADH + H+ so the BPA degradation can continue (Sasaki et al., 2005b). The bisdAB genes from S. bisphenolicum AO1 were isolated, transformed into and expressed in E. coli and enabled this bacterium to degrade BPA, too (Sasaki et al., 2008). In addition, the BisdAB proteins from S. bisphenolicum AO1 were able to degrade BPA in a cell-free system (enzyme assay) in which spinach reductase (EC 1.18.1.2) was added (Sasaki et al., 2005b).
In 2008, the iGEM team from the University of Alberta submitted the bisdAB genes from S. bisphenolicum AO1 to the registry of standard biological parts in the so called Freiburg BioBrick assembly standard (<partinfo>K123000</partinfo> and <partinfo>K123001</partinfo>). In order to degrade BPA in a cell free system, a FNR BioBrick is also needed. As demonstrated, the BisdA and BisdB work together in a cell-free system with spinach reductase and they work intracellular in E. coli, it can be assumed that it is possible to produce all components necessary to degrade BPA in a cell free system in E. coli. To achieve this a ferredoxin-NADP+ oxidoreductase BioBrick, isolated from E. coli, is provided by the Bielefeld-Germany 2011 iGEM team (<partinfo>K525499</partinfo>).
The whole electron transport chain between the three enzymes involved in BPA degradation and the BioBricks needed to enable this reaction in vitro are shown in the following figure (please have some patience, it's an animated .gif file):
Further applications of bisphenol A degrading BioBricks and enzymes
References
Sakai K, Yamanaka H, Moriyoshi K, Ohmoto T, Ohe T (2007) Biodegradation of Bisphenol A and Related Compounds by Sphingomonas sp. Strain BP-7 Isolated from Seawater, Bioscience, Biotechnology, and Biochemistry 71(1):51-57.
Sasaki M, Maki J, Oshiman K, Matsumura Y, Tsuchido T (2005a) Biodegradation of bisphenol A by cells and cell lysate from Sphingomonas sp. strain AO1, Biodegradation 16(5):449-459.
Sasaki M, Akahira A, Oshiman K, Tsuchido T, Matsumura Y (2005b) Purification of Cytochrome P450 and Ferredoxin, Involved in Bisphenol A Degradation, from Sphingomonas sp. Strain AO1, Appl Environ Microbiol 71(12):8024-8030.
Sasaki M, Tsuchido T, Matsumura Y (2008) Molecular cloning and characterization of cytochrome P450 and ferredoxin genes involved in bisphenol A degradation in Sphingomonas bisphenolicum strain AO1, J Appl Microbiol 105(4):1158-1169.
Stolz A (2009) Molecular characteristics of xenobiotic-degrading sphingomonads, Appl Microbiol Biotechnol 81:793-811.
 "
"