Team:UCL London/Research/Supercoiliology/Theory
From 2011.igem.org
| Line 31: | Line 31: | ||
The specific linking difference (σ) is the number of turns added or removed relative to the total number of turns in a relaxed plasmid and thus indicated the level of supercoiling. | The specific linking difference (σ) is the number of turns added or removed relative to the total number of turns in a relaxed plasmid and thus indicated the level of supercoiling. | ||
| + | |||
σ = (Lk-Lk(0))/Lk(0) | σ = (Lk-Lk(0))/Lk(0) | ||
Revision as of 03:18, 22 September 2011
The Theory of Gyrase
DNA gyrase was discovered in 1976 by a group of researchers (Martin Gellert et.al.) working at the National Institute of Health, Bethesda, Maryland, USA [1]. This prokaryotic enzyme is known to be the only one under its class (topoisomerase type II) and is the only topoisomerase capable of introducing negative supercoils into closed circular plasmid molecules. This unique negative supercoiling reaction requires magnesium ions and adenosine triphosphate (ATP) as the energy source. DNA gyrase therefore plays an important role in the growth as well as in the replication of E. coli.
A series of studies have reported that DNA gyrase relaxes the stress which is built up ahead of the gene transcription and DNA replication machinery. The stress of positive supercoils which accumulates downstream of these transcribing and replicating protein complexes is relieved by DNA gyrase molecules to allow for easier and obstruction-free propagation of the participating DNA or RNA polymerase complexes.
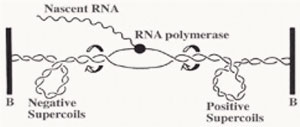
 Furthermore, negatively supercoiled plasmids also enables easier transition from the closed promoter complex to the open promoter complex and this accounts for higher transcriptional efficiency of the underwound B-DNA (right handed helix) molecule [2]. Although in theory, this indicates that all promoters will therefore have better transcriptional efficiency, but as usual there are specific exceptions.
Furthermore, negatively supercoiled plasmids also enables easier transition from the closed promoter complex to the open promoter complex and this accounts for higher transcriptional efficiency of the underwound B-DNA (right handed helix) molecule [2]. Although in theory, this indicates that all promoters will therefore have better transcriptional efficiency, but as usual there are specific exceptions.
In E.coli, the level of supercoiling actually acts as a regulatory mechanism for controlling the transcription and subsequent expression of gyrase molecules inside the cell. Gyrase genes are known to be repressed when the extent of negative supercoiling exceeds a certain threshold level. This molecular phenomenon is due to the spatial conformation adopted by the DNA sequences flanking the gyrase genes and promoters [2]. The transcription of gyrA and gyrB genes, which code for the subunits of DNA gyrase, is induced by relaxation of the DNA molecule. It has been proposed that promoter clearance instead of open complex formation is the actual rate-limiting step for these promoters.
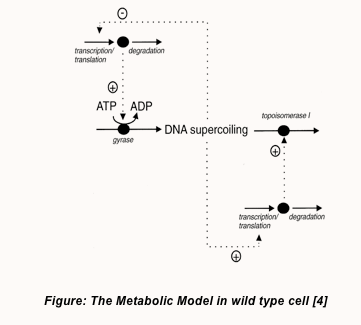
In the presence of a high degree of negative supercoiling in the DNA molecule, the expression of DNA gyrase is seen to repressed while that of topoisomerase I seems to be upregulated automatically. This topoisomerase I is another enzyme which is responsible for the relaxation of negative supercoils in the DNA as opposed to the property of gyrase. The presence of this dual antagonistic enzymes therefore acts as a regulatory mechanism for maintaining a constant superhelical density (σ) and consistent topology within the E. coli chromosome. Under normal physiological conditions optimal to cell growth, the average superhelical density in an E coli cell is equivalent to 0.05 as indicated by the work carried out by Sinden et al. (1980) [3]. A model of this regulatory mechanism is displayed below.
The gyrase expression cassette that we have designed in our project, positions the gyrase subunit ORFs (open reading frames) downstream of the inducible lac promoter (pLac). With this approach, we therefore expect to eliminate the regulatory controls exerted by the wild type gyrase promoters and consequentially the increase in negative supercoiling will no longer restrict gyrase expression. Based on this we can clearly expect an increase in the degree of negative supercoiling of the cellular DNA of a cell which over-expressed gyrase. However, this change in the average superhelical density of the cellular chromosome will affect its growth rate and can possibly produce adverse effects which eventually leads to senescence.
Parameters defining the topology of plasmid DNA
For DNA molecules present within a cell, it is quite common for one strand to be always wound around the other in order to form the DNA double helix. Following this if the DNA molecule is then to become supercoiled, it must form a closed loop with its ends completely fixed. Once its ends are covalently fixed together, it is said to be in its relaxed topological state and can therefore be described by the following equation and parameters [2]:
Lk = Tw + Wr
Lk = Linking number (number of double helical turns in the original linear molecule) Tw = The helical twist (represents the way in which DNA strands coil around each other about the axis of the DNA helix) Wr = superhelical turns (measure of the contortion of the helix axis in space)
In a relaxed DNA plasmid, Lk is defined as Lk(0) and is equal to Tw while Wr=0. Tw= N/h, where h is the number of base pairs per turn of the DNA helix and N is the number of base pairs in the entire DNA molecule.
The specific linking difference (σ) is the number of turns added or removed relative to the total number of turns in a relaxed plasmid and thus indicated the level of supercoiling.
σ = (Lk-Lk(0))/Lk(0)
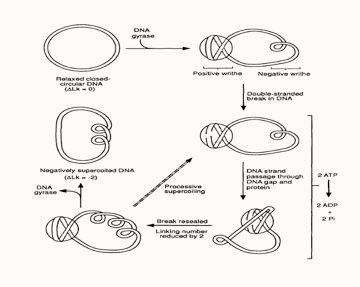
Gyrase action mechanism
Current gyrase activity models, proposes that the tetrameric gyrase complex binds to the gyrase binding site in a DNA molecule to form the gyrase-DNA complex, which comprises of 120bp of DNA sequence wrapped around the protein molecule in the form of a positive supercoil. Since gyrase-DNA complex introduces a positive supercoil into the closed circular plasmid, this gives rise to another negative supercoil (writhe) nearby within the plasmid molecule in order to balance out the positive supercoil, as the linking number of a plasmid always remains constant unless the sequence has a disruption or a ‘nick’.?
Following the association with DNA, gyrA subunit of the enzyme progresses to break the DNA strand and its Tyr122 residue forms a phosphotyrosine linkage with the cleaved 5’ end of the DNA backbone, while the 3’ end is suspended between the enzyme and the DNA by non-covalent forces. Each gyrA subunits form one individual phosphotyrosine linkage with each DNA strand and allows them to pass through the breakage gap, following which the gap is resealed by means of a nucleophilic attack between the oxygen in the 3’ hydroxyl group and the phosphotyrosine linkage on the 5’ end. In this way gyrase reduces the linking number by two in the plasmid molecule and a single cycle of this reaction costing two ATPs consumed by the two gyrB subunits. In the reaction cycle, divalent cations such as Mg2+ ions facilitates binding of the gyrB subunit active sites to the beta and gamma phosphate groups of ATP. Thus, the introduction of a suitable chelating agent into a gyrase reaction mixture produces a reduction in gyrase activity. Additionally, the use of ATP analogues, like AMP-PMP for instance, allows for the visualisation of the gyrB subunits by X-ray crystallography [2]. </html>
 "
"