Team:Paris Liliane Bettencourt/Notebook/2011/07/25/
From 2011.igem.org

Cyrille
The positive transformation controls of the previous QCM where good, but there were no colonies where found on the plates.
The manipulation was attempted again. As a doubt was on the dNTPs quality, the negative control was made with no dNTPs. The protocol is the same than the one of the 23/07.
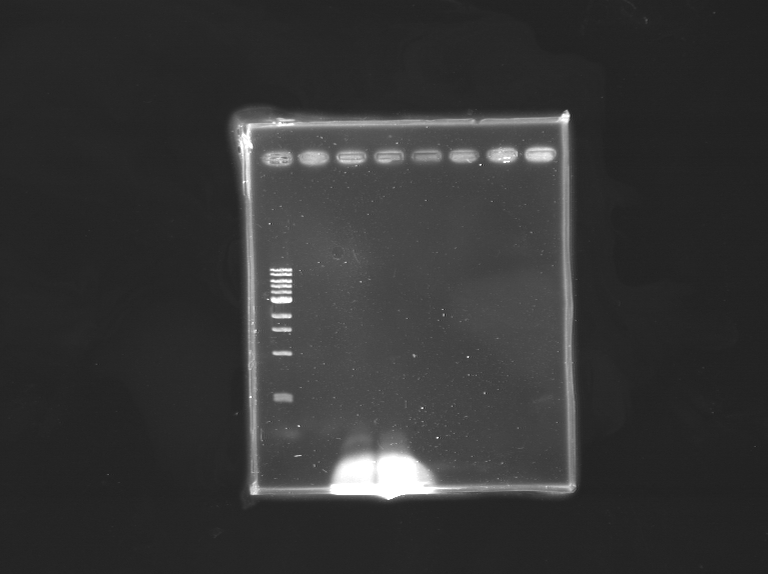
This time a gel was loaded with the samples before and after the digestion by DpnI. The result is that nothing is to be seen on any of the gels, excepts the primers.
The conclusion is that there is nothing as a template in the plasmid tube despite the measure gives a high concentration. The plated transformation gives no results. We notices that the positive transformation control with the turbocells was not very strrong contrary to what was previously obtained with other transformants.
As a doubt was raised on the template DNA tube, we runned a gel with just the plasmid, and the plasmid digested by EcoRI. No band was visible on the gel.
From a glycerol we launched a miniprep. The mini-prep was done by Axel the day after.
Camille & Danyel
We purified the PCR product from the overnight PCR launched the day before and measured the concentration with the Tecan : 3209.2ng/µL with an 260nm/280nm absorbance ratio of 1.73 .
A discussion with our instructor Ariel leads us to do an other PCR with a new protocole :
- 2min at 95°C
- 50s at 60°C (annihiling temperature of our primers)
- 1h at 72°C (elongation temperature of Phusion polymerase)
We mixed in our PCR tubes :
- 0.20µL of forward primer(initial solution concentration : 39µmol/L)
- 0.20µL of Reverse primer(initial solution concentration : 48µmol/L)
- 0.20µL of Phusion polymerase (Themo Scientific)
- 4µL of Phusion buffer
- 0.4µL of dNTPs mix
- 14.4µL of water
We measured the concentration and ratio of the PCR product :
- [DNA]=424.8ng/µL
- 260nm/280nm absorbance ratio = 1.25
For the control :
- [DNA]=103.8ng/µL
- 260nm/280nm absorbance ratio = 1.11
After PCR putification [DNA]=18.3ng/µL in 8µL
Still following the instructions given by our instructor for this protocole we did a digestion of 34.6ng of DNA from this product.
A new PCR putification to get rid of the restriction enzymes led to a [DNA]=16.4ng/µL with an 260nm/280nm absorbance ratio = 2.28.
Our second part of the project the T7 with amber mutation in order to use the tRNA we've just made, needs a QuickChange mutagenesis that we schedule to do tomorrow.
 "
"