June 26
Start overnight cultures of T7 polymerase (K145001), mCherry (J06702), Lac Promoter (R0010), double terminator (B0014 and B0015) and Tet Promoter (R0040)
June 27
Miniprep of T7 polymerase (K145001), mCherry (J06702), Lac Promoter (R0010), double terminator (B0014 and B0015) and Tet Promoter (R0040) and submit for sequencing
Transfer 0.5 mL aliquots of BPA and 5 mL aliquots 17a-ethynylestradiol cultures from June 23 to fresh minimal media
Checked DDT and nonylphenol cultures, will wait until tomorrow to transfer
Ethanol precipitation of DNA extractions from LA river samples following protocol
Prepare antibiotic stocks (Ampicillin, Chloramphenicol)
Results
Miniprep
| Part |
Concentration(ng/ul) |
| B0014 |
230.5 |
| B0015 |
254.1 |
| J06702 |
301.3 |
| K145001 |
205.6 |
| R0010 |
117.3 |
| R0040 |
156.8 |
We found growth in one of our BPA enrichment cultures, and slight growth in our ethinyl estradiol cultures. We transferred these vials today.
June 28
Send off continued forward sequencing for HER
PCR for Gibson assembly of PNT001 and PNT002
Gel and PCR purification of PCR products
Analysis of sequencing results from yesterday
Transfer DDT and nonylphenol cultures to new media
Transform mCherry (J06702), Lac Promoter (R0010), double terminator (B0014 and B0015) and Tet Promoter (R0040) for creation of glycerol stocks
Results
Sequencing: All biobricks showed correct sequence except T7 Polymerase
Ran a gel of the PCR products. Strangely, lanes 3 and 4 show no DNA. We will repeat the PCR of lanes 3-5 and Gibson assemble pNT001 (R0010, K123000, B0014) later in the week.
EtOH precipitation of Soil Extractions from June 21
| Tube/Location Number |
Concentration(ng/ul) |
| 1 |
13.6 |
| 2 |
15.2 |
| 4 |
8.3 |
| 5 |
11.4 |
| 6 |
25.5 |
| 7 |
23.5 |
| 9 |
261.3 |
| 10 |
74.0 |
June 29
Redo PCR of R0010 and K123000
Analyze sequence of ER, design new primer for continued sequencing
Prepare overnight cultures of K145001 colonies for sequencing and overnight cultures of other biobrick plates for creating glycerol stocks
Plan experiments using pNT001 and pNT002
Results
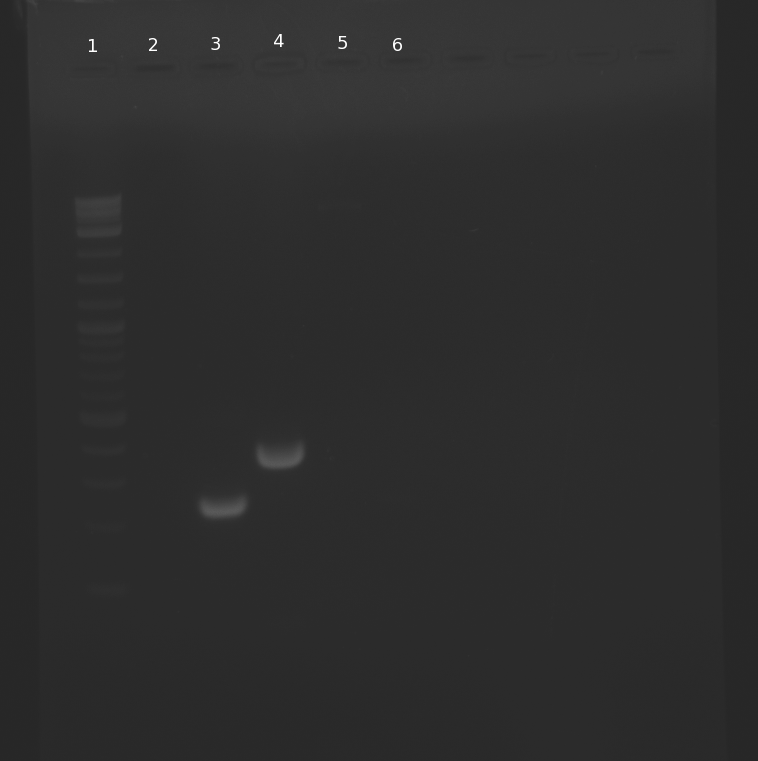
PCR of parts for pNT001(BioBricks+primers for Gibson): 1 ladder; 2 blank; 3
R0010; 4
K123000; 5
B0014; 6 primers from lane 3 for negative control
Estrogen receptor (K123003) sequencing is back. Again, the translation is right, but the bases don't match. The stop codon is not in this read. We will keep sequencing.
B0014 was not amplified in PCR. We will PCR this part and primers again, as the PCR from June 28 has a rather low concentration to be used for Gibson Assembly.
Concentration of Purified PCR products from June 28 and June 29
June 30
Miniprep 5 K145001 cultures and send them off for sequencing
Make glycerol stocks of mCherry (J06702), Lac Promoter (R0010), double terminator (B0014 and B0015) and Tet Promoter (R0040)
Repeat PCR of B0014 and perform PCR to linearize backbone vector (pSB4A5)
Digest PCR products with DpnI and purify
Results
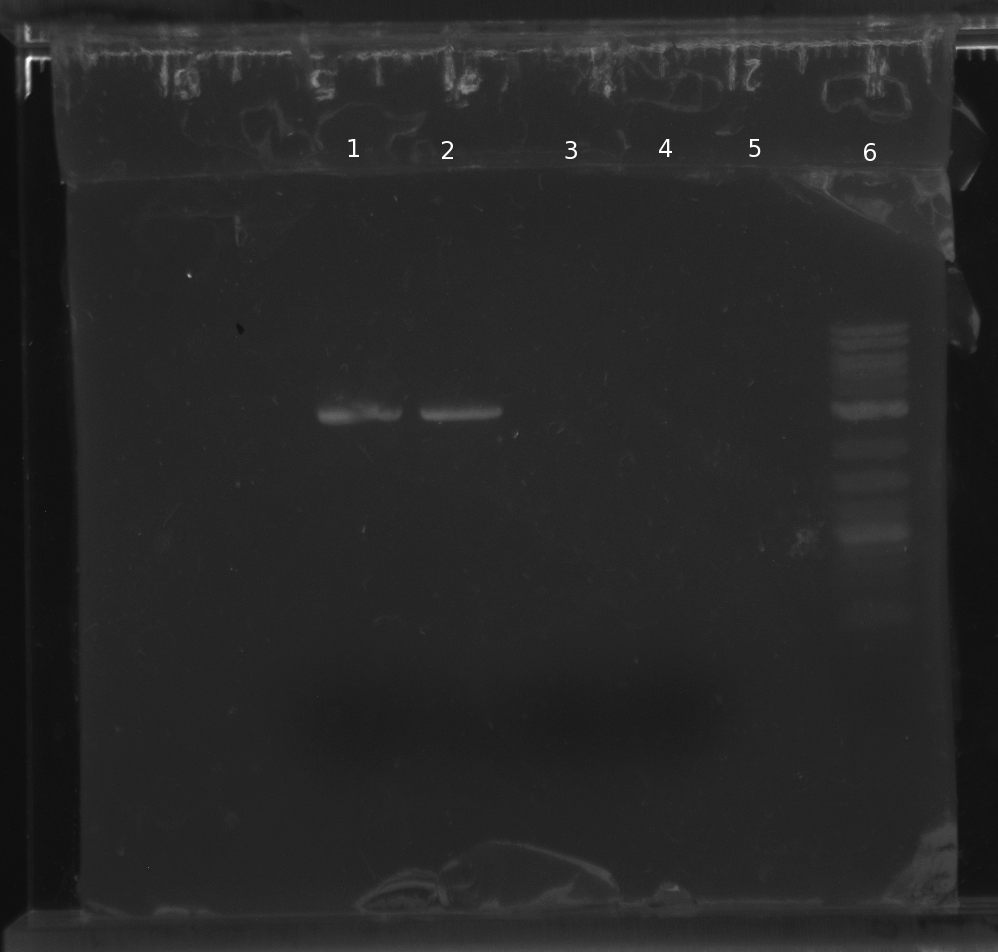
PCR of parts for pNT001 and pNT002: 1 & 2
B0014; 3 & 4
pSB4A5; 5 blank; 6 ladder
B0014 appeared in the gel, but pSB4A5 did not. We are redoing PCR of pSB4A5 with primers to add the prefix and suffix with longer annealing and elongating times.
Concentration of the 5 K145001 minipreps:
| Miniprep |
Concentration (ng/ul) |
| 1 |
154.7 |
| 2 |
356.7 |
| 3 |
153.2 |
| 4 |
146.4 |
| 5 |
138.2 |
July 1
Dpn1 digest and purification of pSB4A5 PCR
Run a gel of pSB4A5 PCR
Check sequencing of K145001
Check if enrichment cultures are ready for transfer
Try running PCR at different annealing temperatures (55˚, 60˚, 65˚ C)
Try the PCR, using the 4x5backbone primers on pSB4K5
Order more 4x5 backbone primer
Transform pSB4A5 from the distribution plate into XL-10 gold competent cells in case our source of plasmid is bad
Results
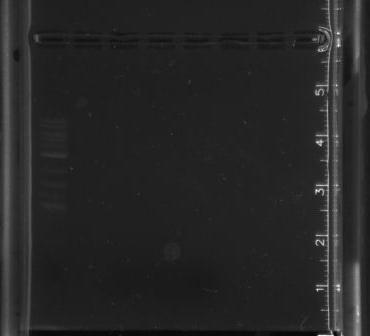
PCR of pSB4A5: 1 ladder; 2 blank; 3, 4, 5 pSB4A5

PCR of 4x5 primers with pSB4A5 and pSB4K5 at different annealing temperatures: 1 ladder; 2 blank; 3 pSB4A5 55˚C; 4 pSB4K5 55˚C; 5 pSB4A5 60˚C; 6 pSB4K5 60˚C; 7 pSB4A5 65˚C; 8 pSB4K5 65˚C
No bands appeared in the first gel (right), indicating that the PCR did not work.
PCR using different annealing temperatures had bands in every experimental lane, indicating that the PCR worked.
Aligned K145001 to sequence reads of the five samples. Sample 2 is most likely one containing the T7 polymerase, as the alignment seems best and BLASTn searches through Geneious of forward and reverse sequence reads both give a hit of NC_001604, part of the T7 genome.
| 6/30 B0014 PCR purification |
Concentration (ng/ul) |
| 1 |
30.3 |
| 2 |
31.5 |
July 2
Gibson assemble pNT001 and pNT002 with negative controls and transform into competent cells
Check if enrichment cultures are ready for transfer
Results
Concentration of pSB4A5 PCRs:
| pSB4A5 PCR temp. (C) |
Concentration (ng/ul) |
| 55 |
73.4 |
| 60 |
127.5 |
| 65 |
83.9 |
All of the enrichment cultures remain clear.
 "
"





