Team:Washington/Parts
From 2011.igem.org
(Difference between revisions)
(→Diesel Production) |
(→Gluten Destruction) |
||
| Line 25: | Line 25: | ||
==='''Gluten Destruction'''=== | ==='''Gluten Destruction'''=== | ||
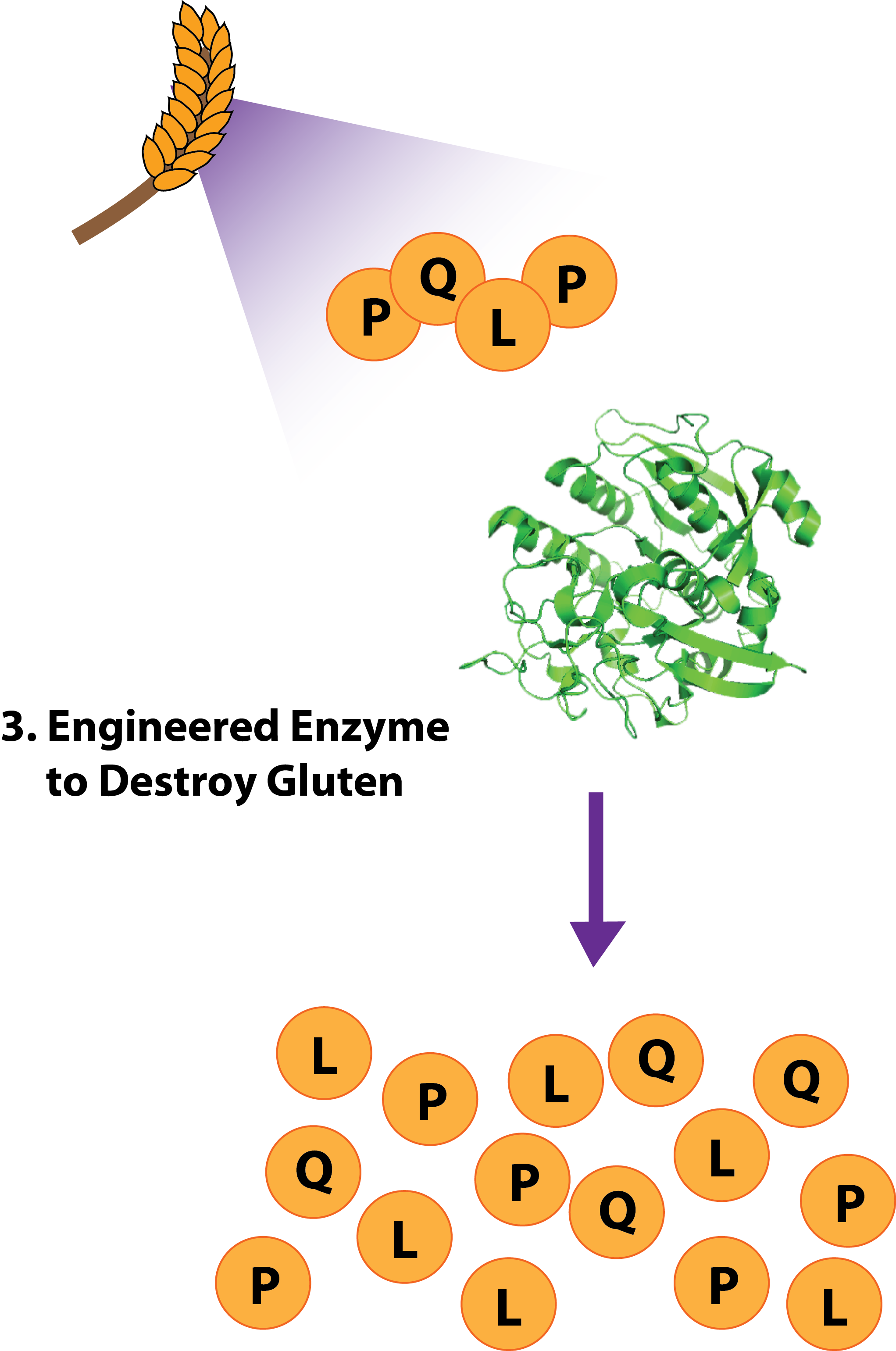
| - | :: 3. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590023 BBa_K590023: '''KumamaMax''']- A modified version of the enzyme Kumamolisin, a protease ofthe sedolisin family native to ''Alicyclobacillus sendaiensis'' known to be active at low pH and elevated temperatures. To Kumamolisin, the mutations N291D, G319S D358G, D368H increase activity to the PQLP peptide, an antigenic epitope in gliadin, 118-fold. | + | :: '''3.''' [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590023 BBa_K590023: '''KumamaMax''']- A modified version of the enzyme Kumamolisin, a protease ofthe sedolisin family native to ''Alicyclobacillus sendaiensis'' known to be active at low pH and elevated temperatures. To Kumamolisin, the mutations N291D, G319S D358G, D368H increase activity to the PQLP peptide, an antigenic epitope in gliadin, 118-fold. |
==='''Gibson Assembly Toolkit'''=== | ==='''Gibson Assembly Toolkit'''=== | ||
Revision as of 04:53, 23 September 2011
Data Summary
Data for Favorite New Parts
Diesel Production
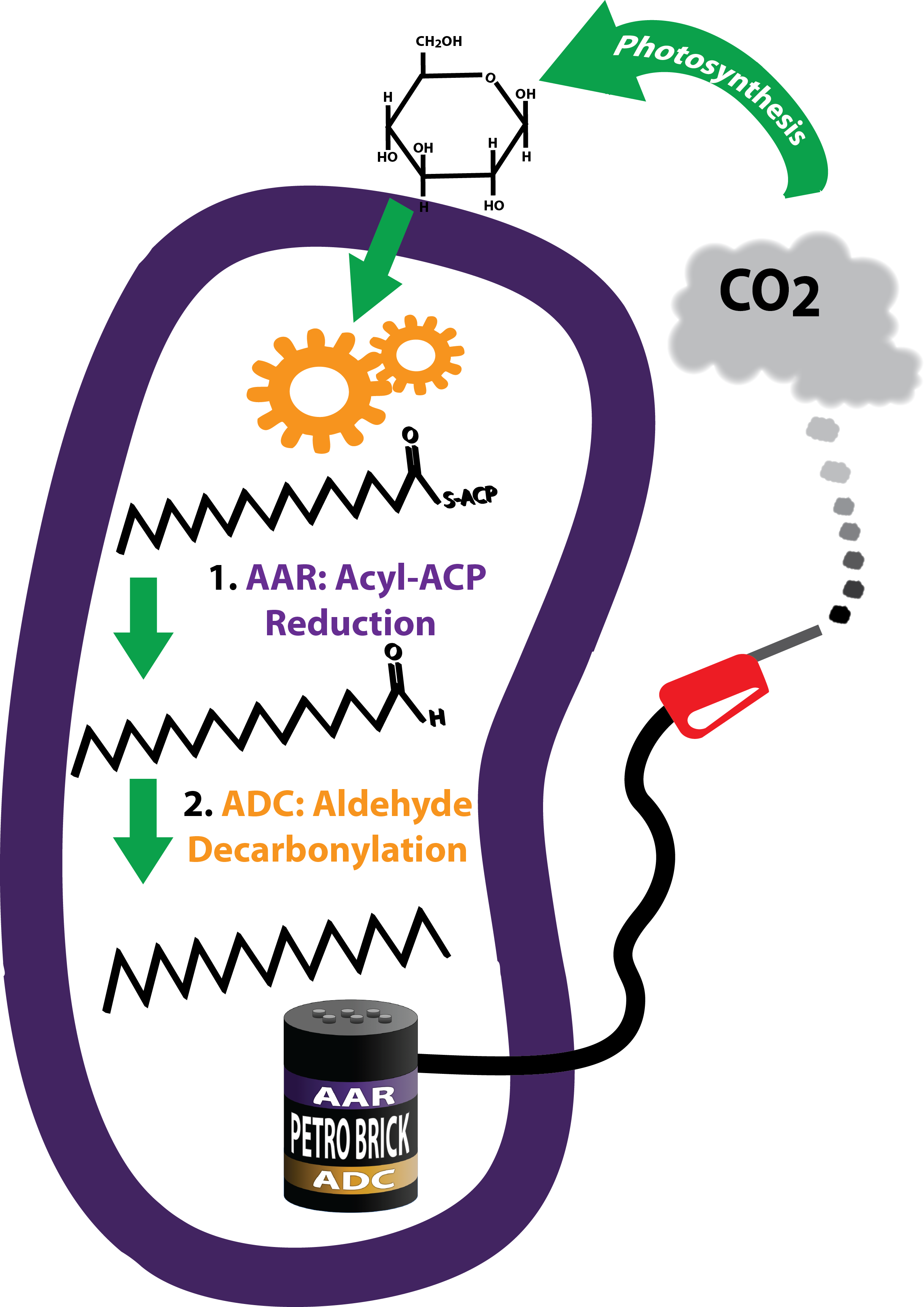
- 1, 2. [http://partsregistry.org/Part:BBa_K590025 BBa_K590025: The PetroBrick] - A modular and open platform for the biological production of diesel fuel. The PetroBrick consists of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590032 AAR] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590031 ADC], each behind a standard Elowitz RBS. All of this is under regulation by a high constitutive promoter in pSB1C3.
Gluten Destruction
- 3. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590023 BBa_K590023: KumamaMax]- A modified version of the enzyme Kumamolisin, a protease ofthe sedolisin family native to Alicyclobacillus sendaiensis known to be active at low pH and elevated temperatures. To Kumamolisin, the mutations N291D, G319S D358G, D368H increase activity to the PQLP peptide, an antigenic epitope in gliadin, 118-fold.
Gibson Assembly Toolkit
- 4. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590010 BBa_K590010: pGA1A3_pLacGFP], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590011 BBa_K590011: pGA1C3_pLacGFP], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590012 BBa_K590012: pGA4C5_pLacGFP], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590013 BBa_K590013: pGA4A5_pLacGFP], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590014 BBa_K590014: pGA3K3_pLacGFP] - These are plasmid backbones optimized for use in Gibson cloning, with a variety of copy numbers and antibiotic resistances.
Magnetosome Toolkit
- 5. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590015 BBa_K590015: sfGFP_mamK_pGA1C3] - This part consists of the mamK gene from Magnetospirillum magneticum strain AMB-1, fused to sfGFP, in the backbone of pGA1C3. MamK was previously reported to be required for proper alignment of magnetosomes in a chain in magnetic bacteria.
- 6. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590016 BBa_K590016 sfGFP_mamI_pGA1C3] - This part consists of mamI gene from Magnetospirillum magneticum strain AMB-1, sfGFP in the backbone of pGA1C3. MamI is a membrane-localized protein that localizes magnetic vesicles to the surface of cells, thus forming characteristic magnetosome chains.
Data for Existing Parts
Fill Me In
- [http://partsregistry.org/Part:BBa_J45119:Experience Experience] - Wintergreen odor enzyme generator, BBa_J45119 (MIT, iGEM 2006): 98 out of 100 volunteer subjects standing up to 5 feet away from the bacterial cultures could distinguish wintergreen-producing bacteria from negative controls.
- [http://partsregistry.org/Part:BBa_J61110:Experience Experience] - RBS, BBa_J61110 (Arkin Lab, 2007): Of the 5 RBS Parts we tested, this RBS works best for expressing yellow fluorescent protein-tagged BAR
Improved Parts
Fill Me In
- [http://partsregistry.org/Part:BBa_XXXXX Main Page] - Air Freshilizor, BBa_XXXXX: Our mathematical model predicts that the threshold of activation is 10 parts per billion, the concentration of Butanethiol that humans can typically smell
 "
"