Team:Washington/Alkanes/Future/Localization
From 2011.igem.org
(→Direct Fusion) |
(→Localization) |
||
| Line 20: | Line 20: | ||
| - | + | ||
==Zinc Finger Fusion== | ==Zinc Finger Fusion== | ||
Revision as of 01:43, 23 September 2011
Localization
To improve the efficiency of acyl-ACP conversion through the two-step pathway to alkane production, we are performing optimization through localization of the two catalytic proteins. Decrease of the distance the intermediates must travel between Acyl-ACP Reductase (AAR) and Aldehyde Decarbonylase (ADC) is expected to improve overall alkane synthesis.
Initially, we attempted a direct fusion of the two enzymes with a linker. Our main approach is to optimize alkane production by re-use of Biobrick parts from a previous iGEM team. For this, we are re-adapting Biobrick parts from 2010 Slovenia's zinc finger violacein biosysnthesis project. By colocalization of the individual violacein producing enzymes in it's five-step pathway, they were able to increase their pigment production by ~6-fold. Our goal for alkane production optimization is to use the zinc finger localization method to significantly increase yields.
Direct Fusion
Our first approach was direct fusion of two proteins using a glycine-serine linker. We chose to construct both of the linear configurations of AAR to ADC linkage to test the effects of fusion by N and C terminus regions on either enzyme's catalytic performance. This resulted in both the AAR-to-ADC and ADC-to-AAR fusion products.
<partinfo>BBa_K590051 DeepComponents</partinfo>
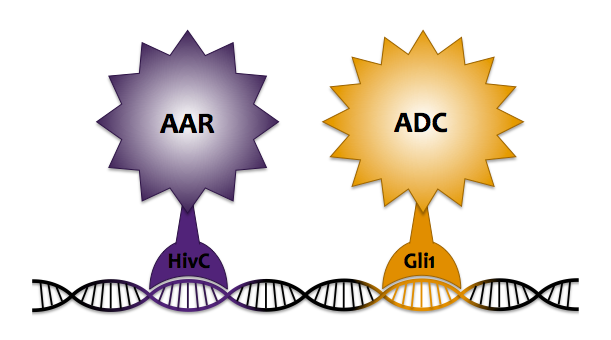
Zinc Finger Fusion
In an attempt to enhance our localization technique beyond the simple protein fusion method, we chose to integrate the use of DNA localization via zinc finger proteins to colocalize ADC and AAR to increase system flux. Zinc fingers are small DNA binding domains, consisting of around 30 amino acid residues folded around a single zinc cation. They function as transcriptional factors within eukaryotes, regulating endogenous gene expression by binding to specific sequences along DNA. Last year's Slovenia iGEM team characterized 6 zinc fingers and created a DNA program containing specific binding sites for each zinc finger. Our goal was to fuse AAR and ADC to their own respective zinc finger, such that when co-expressed with the DNA program, the fusion proteins would each bind to their specific target sequence along the DNA, thus being held in close proximity so that alkane production could be carried out more efficiently.
 "
"