Results
The strains transformed with the caretenogenic genes are a bright orange color due to all the beta-carotene they produce, as beta-carotene is an orange pigment. We confirmed its presence via HPLC using a standard.
On a first visual comparison of growth on dough media vs YPD we did see a significantly slower growth on dough media than on YPD. This is to be expected considering there is a large excess of nutrients on the YPD plates. There was still significant growth on the dough media plates however, enough to show that it would be viable to use in bread.
We then took it one step further and showed that baking bread with the Vitamin A yeast was possible.
Spectroscopy and HPLC
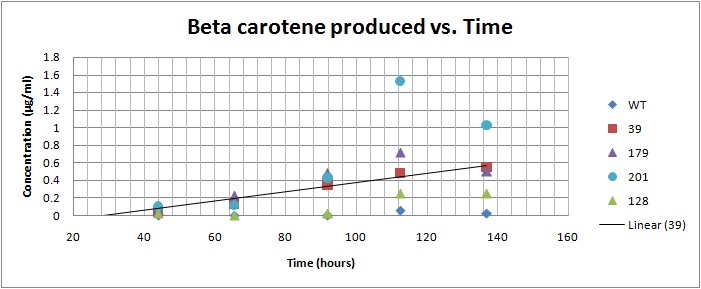
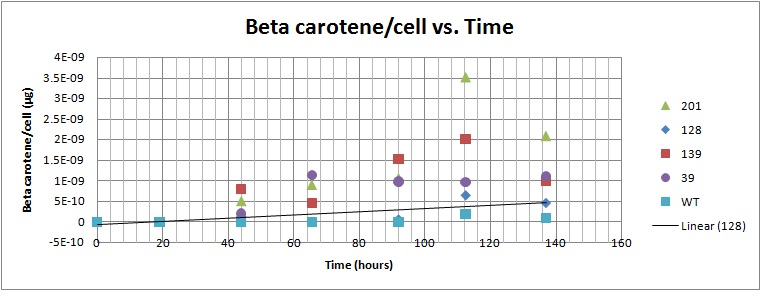
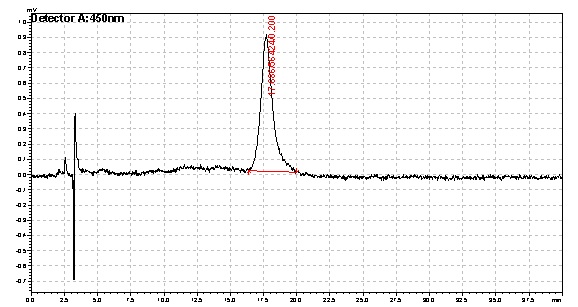
Based on the results of our Spectroscopy experiments measuring beta-carotene versus time and characterization by the use of HPLC, we can see that the production of beta-carotene over time increases at a linear rate. The results of the HPLC confirm that the yeast cells are in fact producing beta carotene, as the absorption peak at 450nm corresponds to the absorption maxima of beta-carotene. Before running the samples on the HPLC, beta carotene was extracted from cell extract using hexane, as it would only isolate highly hydrophobic molecules, such as beta-carotene. As seen in following Spectroscopy and HPLC graphs, beta carotene production and characterization can be quantified. In addition, beta carotene per cell over time can be calculated, which provides initial conditions for our model used to optimize the beta carotene pathway. As seen in the HPLC absorbence spectra on the left, beta carotene shows a strong signal at 450nm, matching beta carotene characterization in literature, allowing us to confirm that the orange color produced in our strains can be attributed to increased beta carotene production.
Protein Purification
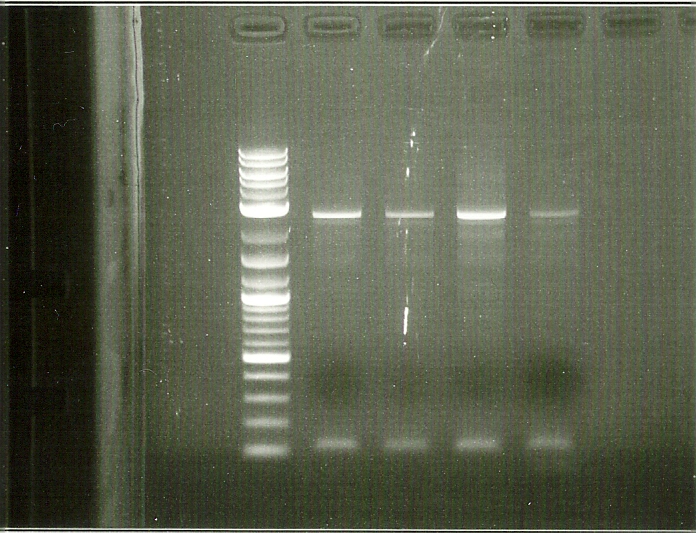
In order to perform enzyme kinetics assays, we needed to first purify our proteins of interest. From the beta carotene pathway, these enzymes were geranylgeranyl pyrophosphate synthase, phytoene desaturase, and a bifunctional phytoene synthase. Using a TEV-his6X-mCherry-KAN tag, we would be able to purify our enzyme with only one high efficiency purification step (Nickel Column Purification using His-tag) and characterize product with great accuracy through the use of anti-mCherry antibodies on the RFP component of the tag. The goal of the PCR was to first produce a PCR product that contained the HIS-TEV-mCherry-KAN-CYC1tt tag. Another PCR would enable us to tag each of our genes of interest, which we could then transform into yeast for expression and purification. As depicted in the PCR gel to the left, both rounds of PCR proved to be successful, as we obtained a large signal for each of our final tagged genes. These results enabled us to transform these tagged genes into yeast.
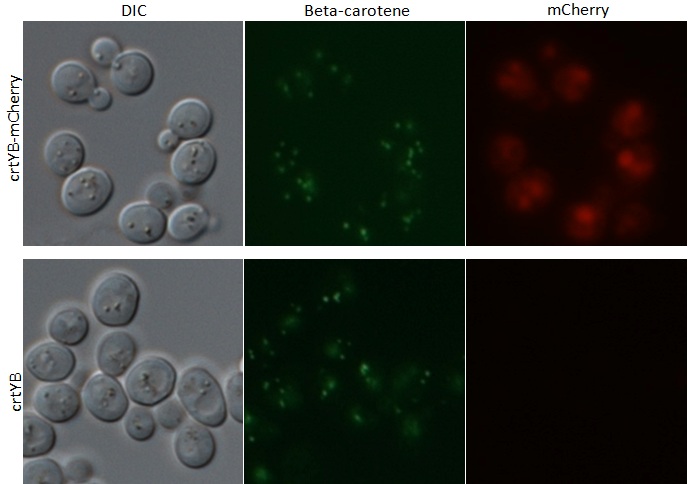
After transformation of our tagged genes, fluorescence microscopy was used to success of the transformation and expression of our tagged genes. Depicted in the fluorescence microscopy figure, the transformed yeast cells fluorescence red, which is evidence that our tagged mCherry (RFP) tagged genes are in fact present and being expressed. Green fluorescence is seen in both wild type and transformed yeast, which is due to the presence of beta carotene.
References
1. High-Level Production of Beta-Carotene in Saccharomyces cerevisiae by Successive Transformation with Carotenogenic Genes from Xanthophyllomyces dendrorhous René Verwaal,1, Jing Wang,1 Jean-Paul Meijnen,1 Hans Visser,1,Gerhard Sandmann,2 Johan A. van den Berg,1, and Albert J. J. van Ooyen1*
 "
"