|
|
| (17 intermediate revisions not shown) |
| Line 5: |
Line 5: |
| | Related Links:</div> | | Related Links:</div> |
| | <div id="boxcontent"><DL> | | <div id="boxcontent"><DL> |
| - | <div class="heading">Vitamin C:</div> | + | <div class="heading">On This Page:</div><DL> |
| - | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Team/Modeling/VitC">Modeling</a><br/> | + | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Project/VitA#Relevant_Parts">Relevant Parts</a><br/></DL> |
| - | <DD><a href="#">Parts</a><br/></br/></DL>
| + | <div class="heading">Vitamins:</div><DL> |
| - | <div class="heading">Projects:</div><DL> | + | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Vit/Bg">Background</a><br/> |
| - | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Team/Project/PromUTR">Promoters and UTRs</a><br/> | + | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Vit/Over">Overview</a><br/> |
| - | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Project/Violacein">Violacein</a><br/> | + | |
| | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Project/VitC">Vitamin C</a><br/> | | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Project/VitC">Vitamin C</a><br/> |
| - | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Project/Vector">Yeast Vector Library</a>
| |
| | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Project/MeasureQuant">Measurements</a> | | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Project/MeasureQuant">Measurements</a> |
| | + | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Project/Baking">Applications</a><br/> |
| | + | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Vit/Results">Results</a><br/> |
| | + | <DD><a href="https://2011.igem.org/Team:Johns_Hopkins/Vit/Future">Future Plans</a><br/> |
| | </DL> | | </DL> |
| | </div> | | </div> |
| Line 26: |
Line 27: |
| | [[File:VitA2.png|610px|betacarotene]] | | [[File:VitA2.png|610px|betacarotene]] |
| | '''''beta''-Carotene''' | | '''''beta''-Carotene''' |
| - |
| |
| - | ======Metabolic engineering======
| |
| - |
| |
| - | ''S. cerevisiae'' has endogenous machinery to produce farnesyl diphosphate, but the rest of the pathway must be engineered. See this schematic for a complete view of the synthetic pathway.
| |
| - |
| |
| - | Previous work has been conducted in which the successful implementation of the Beta-carotene producing enzymes have been engineered into ''S. cerevisiae'' from carotenogenic genes from X. dendrorhous. This pathway can be viewed here1.
| |
| | | | |
| | ====== Pathway ====== | | ====== Pathway ====== |
| - | The Vitamin A production pathway we used as a template to move into Saccharomyces cerevisiae is from another strain of yeast Xanthophyllomyces dendrorhous which does produce beta carotene. S. cerevisiae has endogenous machinery to produce farnesyl diphosphate, but the rest of the pathway must be engineered. The caretenogenic genes that needed to be moved over were carotene desaturase, GGPP synthase, phytoene synthase. The pathway for its production is as follows: | + | The Vitamin A production pathway template integrated into ''Saccharomyces cerevisiae'' was obtained from another strain of ''beta''-Carotene-producing yeast, ''Xanthophyllomyces dendrorhous''. ''S. cerevisiae'' has endogenous machinery to produce farnesyl diphosphate, but the rest of the pathway must be engineered. The missing caretenogenic genes included carotene desaturase, GGPP synthase, and phytoene synthase. The pathway for Vitamin A production is as follows: |
| | | | |
| | [[File:Beta-carotene schematic (1).jpg|610px]] | | [[File:Beta-carotene schematic (1).jpg|610px]] |
| | | | |
| - | Once we had the strains with the genes in them on expression cassettes we verified betacarotene production using HPLC analysis. Being sure that the cells were indeed producing betacarotene, we tested its efficiency on a new kind of media, 'dough media'. Dough media is a watered down version of dough with agar. As the final substrate we wanted to use our yeast on was dough we wanted to see how well it performed. We characterized its beta carotene production over time both on YPD plates and on our dough media plates to see what effect the change of substrate would have on the betacarotene yield from the cells.
| + | To make sure that the cells were indeed producing ''beta''-Carotene, we verified ''beta''-Carotene production using HPLC analysis. Once we were sure the strains containing the missing genes were expressing ''beta''-Carotene, we tested its efficiency on a watered-down combination of dough, our intended final substrate, and agar. We characterized the yeast's ''beta''-Carotene absorption over time on both YPD and dough media plates to observe the effects of substrate change on the ''beta''-Carotene yield from the cells. |
| - | | + | |
| - | Sauer, M., Branduardi, P., Valli, M., & Porro, D. (2004). Production of L-ascorbic acid by metabolically engineered Saccharomyces cerevisiae and Zygosaccharomyces bailii. Applied and environmental microbiology, 70(10), 6086-91. doi: 10.1128/AEM.70.10.6086-6091.2004.
| + | |
| - | | + | |
| - | ======Experiments======
| + | |
| - | | + | |
| - | Vitamin C concentration Assay (By Anne Marie Helmenstine, Ph.D)
| + | |
| - | | + | |
| - | 1. Add 25.00 ml of vitamin C standard solution to a 125 ml Erlenmeyer flask.
| + | |
| - | | + | |
| - | 2. Add 10 drops of 1% starch solution.
| + | |
| - | | + | |
| - | 3. Rinse your buret with a small volume of the iodine solution and then fill it. Record the initial volume.
| + | |
| - | | + | |
| - | 4. Titrate the solution until the endpoint is reached. This will be when you see the first sign of blue color that persists after 20 seconds of swirling the solution.
| + | |
| - | | + | |
| - | 5. Record the final volume of iodine solution. The volume that was required is the starting volume minus the final volume.
| + | |
| - | | + | |
| - | 6. Repeat the titration at least twice more. The results should agree within 0.1 ml.
| + | |
| | | | |
| - | 7. You titrate samples exactly the same as you did your standard. Record the initial and final volume of iodine solution required to produce the color change at the endpoint.
| + | René Verwaal, Jing Wang, Jean-Paul Meijnen, Hans Visser, Gerhard Sandmann, Johan A. van den Berg, and Albert J. J. van Ooyen |
| | + | High-Level Production of Beta-Carotene in Saccharomyces cerevisiae by Successive Transformation with Carotenogenic Genes from Xanthophyllomyces dendrorhous |
| | + | Appl. Envir. Microbiol., July 1, 2007; 73: 4342 - 4350. |
| | | | |
| | + | ======Relevant Parts====== |
| | + | * [http://partsregistry.org/wiki/index.php?title=Part:BBa_K530001 crtE] |
| | + | * [http://partsregistry.org/wiki/index.php?title=Part:BBa_K530000 crtYB] |
| | + | * [http://partsregistry.org/wiki/index.php?title=Part:BBa_K530002 crtI] |
| | | | |
| | | | |
| | <html> | | <html> |
| | </div> | | </div> |
Vitamin A
beta-Carotene (Vitamin A) and other carotenoids are present in many plants and share a common metabolic precursor with critical compounds including chlorophyll, tocopherol (Vitamin E), phylloquinone (Vitamin K1), squalene, and other sterols. Humans cannot naturally produce beta-Carotene, but require it for color vision.
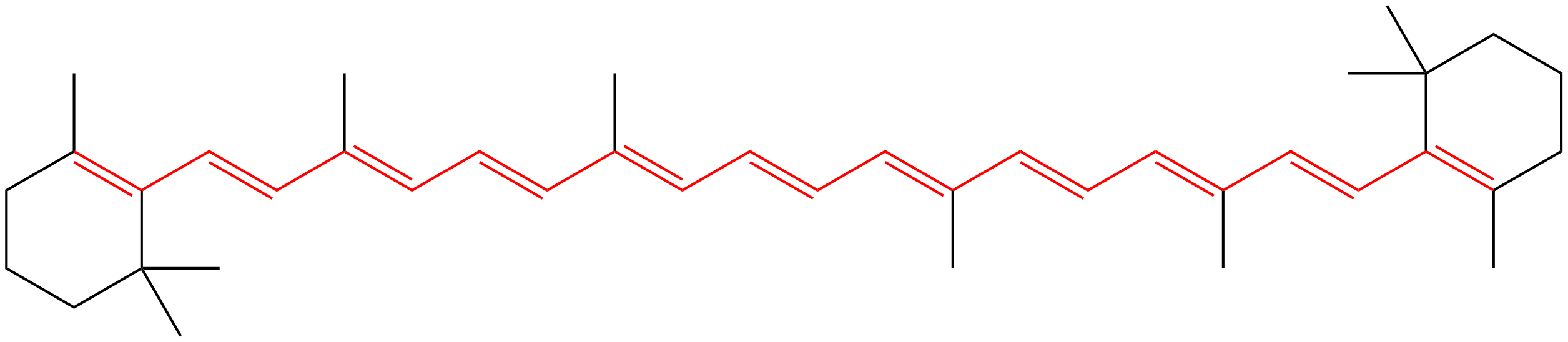 beta-Carotene
beta-Carotene
Pathway
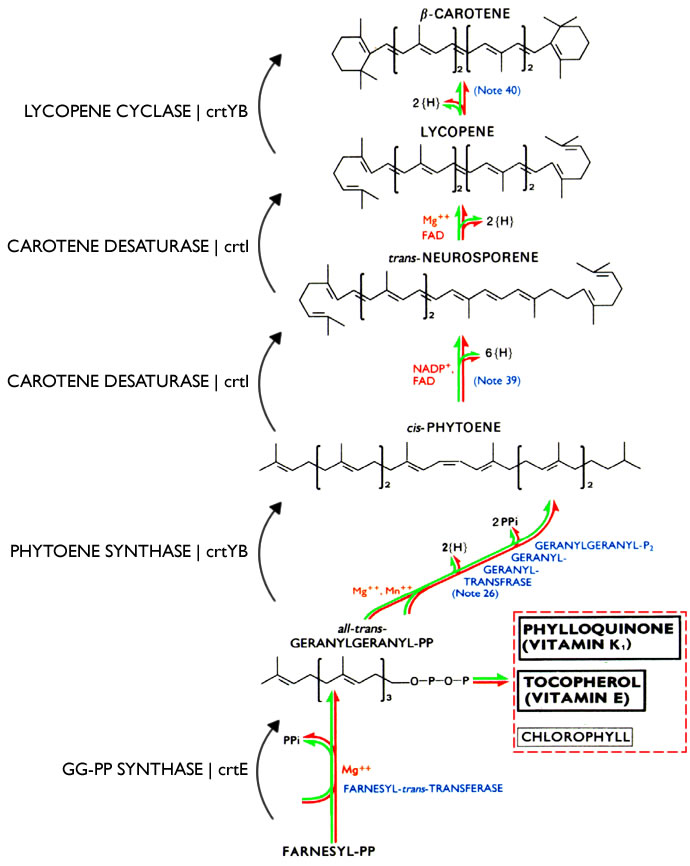
The Vitamin A production pathway template integrated into Saccharomyces cerevisiae was obtained from another strain of beta-Carotene-producing yeast, Xanthophyllomyces dendrorhous. S. cerevisiae has endogenous machinery to produce farnesyl diphosphate, but the rest of the pathway must be engineered. The missing caretenogenic genes included carotene desaturase, GGPP synthase, and phytoene synthase. The pathway for Vitamin A production is as follows:
To make sure that the cells were indeed producing beta-Carotene, we verified beta-Carotene production using HPLC analysis. Once we were sure the strains containing the missing genes were expressing beta-Carotene, we tested its efficiency on a watered-down combination of dough, our intended final substrate, and agar. We characterized the yeast's beta-Carotene absorption over time on both YPD and dough media plates to observe the effects of substrate change on the beta-Carotene yield from the cells.
René Verwaal, Jing Wang, Jean-Paul Meijnen, Hans Visser, Gerhard Sandmann, Johan A. van den Berg, and Albert J. J. van Ooyen
High-Level Production of Beta-Carotene in Saccharomyces cerevisiae by Successive Transformation with Carotenogenic Genes from Xanthophyllomyces dendrorhous
Appl. Envir. Microbiol., July 1, 2007; 73: 4342 - 4350.
Relevant Parts
 "
"



