Creating the Library of Promoters and UTRS
Characterizing the Promoters and UTRs
Measuring the Activity of BioBrick Promoters Using an In Vivo Reference Standard (published in 2009).
9/9/2011
Reference standards received from Imperial College London
Plan: To clone the vectors in E. coli using the Amp selection marker, then miniprep, cut at StuI and transform into yeast.
9/10/11
Standards from Imperial College London transformed into E. coli (protocol specified in the Protocol section).
Cells with plasmids with promoters are plated on LB+Carb plate, and incubated at 37oC overnight.
About 50µL of liquid culture taken from Daniel Wolozny for following inserts:
- Bap2p
- TDH3p
- PRY1p
- KRE9p
- HHO1P
- STM1P
9/11/11
Plates incubated on the previous day are taken out. Colonies grew on the plates for the reference standards. Lawn growth resulted on the the plates for the inserts.
Decide to keep on with the Lawn growth plates anyway; the cultures used to grow the plates were grown from a single colony, so all the cells should be identical.
Take a colony from each plate (or a small section of the plate) into 5 mL of LB+Carb.
9/12/11
Miniprep the grown cultures.
The grown cultures were miniprepped via Zyppy Plasmid Miniprep protocol (Zymo Research).
Concentrations was determined by nanodrop.
9/14/11
Digest by EcoRI and SpeI enzyme, with protocol as specified in the protocol page in the BioBrick protocol.
The provided plasmids from Imperial College London with the reference promoters were digested with StuI enzyme.
Digestions done at 37oC for one hour.
9/16/11
A gel was ran on the digested products to check for products. Products confirmed. One of the three plasmids provided by Imperial College London showed complete digestion.
Proceed to Gel extraction using Qiagen gel extraction kit.
9/18/11
Yeast (YPH500) inoculated for transformation
9/19/11
Yeast transformation:
the StuI digested reference vectors are transformed into the YPH500 culture via protocol specified in the protocol page.
Plated at two concentration of 50 and 350 µL per plate.
Ligation also done for the inserts into the cyc1p EcoRI and SpeI digested plasmids (specific calculation will be included once
9/23/11
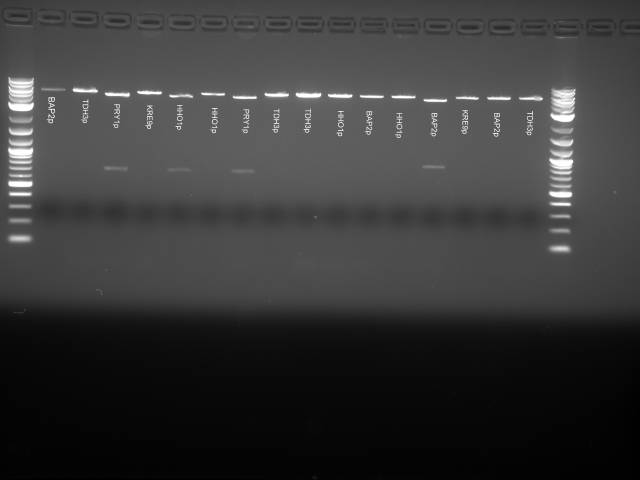
(Second digestion with EcoRI and SpeI are done by Daniel Wolozny and Omar Hadzipasic) A gel is done on the digested samples for verification
This gel shows odd results. The only one with the right size band is the BAP2p plasmid. All the bands in the gel seemed linearized, so it is possible that the plasmids were only cut once.
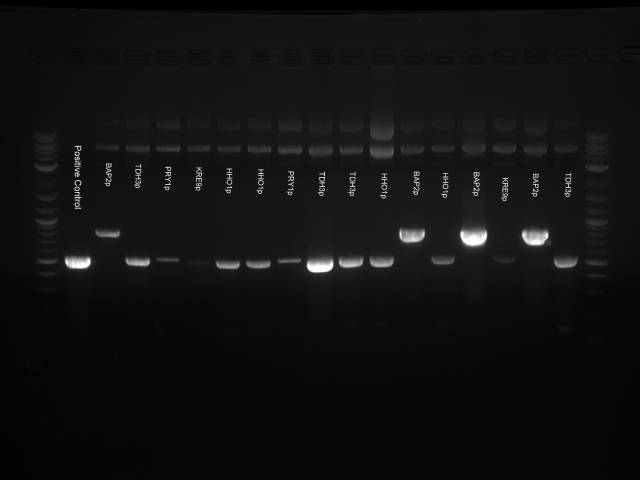
We decided to run a PCR with the primers for the inserts to check if the inserts were present.
PCR with GoTaq Green (Promega):
| Ingredient | Volume (µL) for each one reaction |
| GoTaq Mix | 12.5 |
| Primer mix (10nM) | 2.5 |
| DNA Template | 2 |
| Nuclease Free water | 8 |
 "
"