Team:Imperial College London/Project/Chemotaxis/Overview
From 2011.igem.org
| Line 26: | Line 26: | ||
[[File:ICL_chemotaxis_genetic_constructs.jpg]] | [[File:ICL_chemotaxis_genetic_constructs.jpg]] | ||
<html> | <html> | ||
| - | + | <p>Genetic constructs for expression of malate chemoreceptor are not complicated, however they are modular. Expression of the constructs is under constitutive expression of promoter J23100. It is provided from the registry in the biobrick K398500, in the backbone vector pSB1C3. The coding sequence has been removed from this construct using PCR, providing us with backbone vector with the constitutive promoter, to be assembled using Gibson or CPEC assembly. Ribosome binding sites (RBS) have been generated using Salis RBS calculator with relative translation initiation rates (TIR): 44050 & 42010 for mcpS and PA2652 respectively. The coding sequences for mcpS and for PA2652 have been codon optimised using our codon optimisation software, so that sequence is codon optimised for E. coli and B. subtilis. However we do realise that B. subtilis is Gram-positive bacterium and therefore is not compatible with mcpS and PA2652 receptors. We exploit terminator embedded in the backbone vector pSB1C3 for termination of transcription. In addition, modularity of our system is ensured by putting 15bp insulator sequence between the RBS and the promoter. This sequence was specifically designed to make the promoter interchangeable without affecting the RBS strength and therefore translation initation rate is not affected by changing a different promoter in front of the insulator sequence. This allows us to change the promoter strength simply by changing promoter without modifying the whole genetic construct, thus it improves modularity of the construct. The insulator sequence acts as a starting point for PCR and it can be used to amplify the receptor sequence without the promoter.</p> | |
| - | <p>Genetic constructs for expression of malate chemoreceptor are not complicated, however they are modular. Expression of the constructs is under constitutive expression of promoter J23100. It is provided from the registry in the biobrick K398500, in the backbone vector pSB1C3. The coding sequence has been removed from this construct using PCR, providing us with backbone vector with the constitutive promoter, to be assembled using Gibson or CPEC assembly. Ribosome binding sites (RBS) have been generated using Salis RBS calculator with relative translation initiation rates (TIR): 44050 & 42010 for mcpS and PA2652 respectively. The coding sequences for mcpS and for PA2652 have been codon optimised using our codon optimisation software, so that sequence is codon optimised for E. coli and B. subtilis. However we do realise that B. subtilis is Gram-positive bacterium and therefore is not compatible with mcpS and PA2652 receptors. We exploit terminator embedded in the backbone vector | + | |
| - | + | ||
<p>Root uptake of both <i>E. coli</i> and the bakers’s yeast <i>S. cerevisiae</i> can be observed in the model organism <i>Arabidopsis thaliana</i> and in tomato plants. It is a process that occurs naturally (althought it yet remains to be observed in soil) and we do not need to incorporate additional genes into our design.</p> | <p>Root uptake of both <i>E. coli</i> and the bakers’s yeast <i>S. cerevisiae</i> can be observed in the model organism <i>Arabidopsis thaliana</i> and in tomato plants. It is a process that occurs naturally (althought it yet remains to be observed in soil) and we do not need to incorporate additional genes into our design.</p> | ||
Revision as of 20:02, 29 August 2011
Chemotaxis
Movement performed by bacteria based on attraction or repulsion of chemicals is known as chemotaxis. In our project we are using this mechanism for translocation to plant roots of our modified microbes. They will be attracted to the root and will actively swim towards it.
Our primary chassis for wet lab experiments is Escherichia coli. Chemotaxis in E. coli is well documented. These bacteria can perform two types of movement, tumbling and smooth swimming. The difference between the two is determined by flagellar movement. During tumbling movement, the flagella move clockwise. This is caused by the formation of a complex between CheY-P and FliM, one of the flagella-associated proteins. During smooth swimming, the flagella move counter-clockwise. CheY is not phosphorylated and therefore cannot associate with flagellar proteins, causing the flagella to rotate in the opposite direction.
Smooth swimming is the movement performed by bacteria towards an attractant or away from a repellent. Smooth swimming is controlled by a number of chemotaxis proteins that make up a signalling pathway, with basic functioning having same as typical prokaryotic two component system. First part of the mechanism is sensory kinase, which consists of input domain and autokinase domain. Second part of the mechanism is the response regulator, with reciever and output domains. In the case of chemotactic system, sensory kinase is chemoreceptor associated with CheA and CheW proteins. This association remains present only in the absence of a ligand. During that period CheA autophosphorylates and is capable of phosphorylating CheY, protein which acts as a response regulator in this mechanism. Phosphorylated CheY has the capability of associating itself with flagellar proteins, thereby controlling the direction which flagellum rotates. However, in the presence of ligand, sensory kinase domain is not functional due to dissociation of CheA from chemoreceptor. This way CheY does nto associate with flagellar proteins and result is counterclockwise flagellar movement (Sourjik & Armitage, 2010).
In E. coli chemotaxis there is a number of other proteins, which have functions associated with the two component system and as a result it enables bacterium to move up or down a concentration gradient. This is mediated by CheR, a methyltransferase that methylates MCP (methyl accepting chemotaxis protein). This affects the receptor’s ability to associate with CheW and CheA. Dissociation of CheW and CheA from the chemoreceptor depends on the rising concentration of attractant, which in turn depends on the bacterium moving towards the source of attraction. This is driven by CheZ, a phosphatase that removes phosphate groups from CheY, while sensory kinase is dissociated. In addition, CheB acts as a methylesterase and can remove methyl groups from the MCP receptor, to act as some kind of memory reset (Chelsky & Dahlquist, 1980).
INSERT DIAGRAMSpecification
The chemotaxis module is responsible for ensuring that our bacteria move towards roots. For this, the bacteria need to be able to sense a common root exudate. We have chosen E. coli chemotaxis to be rewired towards malic acid (also referred to as malate), compound found in TCA cycle, which is at low concentrations released form the roots. Since E. coli, the chassis we are using for lab experiments, does not normally exhibit chemotaxis towards malate, we needed to engineer a malate-responsive sensor into the microbes that will enable them to perform chemotaxis towards roots.
Following chemotaxis towards the roots, our bacteria should be taken up into the roots. We want the bacteria to get taken up into the plant roots to ensure that the concentration of indole-3-acetic acid in the plant is increased. If the bacteria remained outside the roots, this goal may also be reached but the risk that we would not increase the internal IAA concentration would be significantly higher. In addition, uptake of bacteria into the roots followed by secretion of chemicals presents a novel platform for modifying plants without engineering their genomes. In a paper published last year, Paungfoo-Lonhienne et al. showed that Arabidopsis and tomato plants are able to actively break down their cell wall to take up GFP-tagged E. coli and S. cerevisiae and use them as a source of nutrients.
Design
While malate-responsive sensors do not occur in E. coli, they have been identified in several other bacteria species, including the soil microbe Pseudomonas putida. We therefore chose to engineer the chemoreceptor mcpS from the P. putida KT2440 strain. McpS is a receptor that responds to a number of TCA cycle intermediates such as malate, fumarate, oxaloacetate, succinate, citrate, isocitrate and butyrate. (Lacal et al, 2010). P. putida and E. coli have different functioning of chemotaxis system, however with structurally similar chemotaxis proteins. Since these proteins are structurally similar, it is reasonable to assume that the mcpS domain will interact with the Che proteins in E. coli and the bacteria will be able to perform chemotactic response upon malate binding to an introduced receptor mcpS. In addition to usage of mcpS chemoreceptor, we are also introducing another malate responsive chemoreceptor PA2652 from the Pseudomonas aeruginosa PA01 strain (Alvarez-Ortega & Harwood, 2007). With PA2652 we are applying the same logic as with mcpS, in that structural similarity is enough for these proteins to be compatible with native chemotaxis system of E. coli. The introduction of two different malate chemoreceptors to different cells, allows us to compare different responsiveness of rewired chemotaxis based on structural similarity of introduced receptors to the common E. coli receptors.
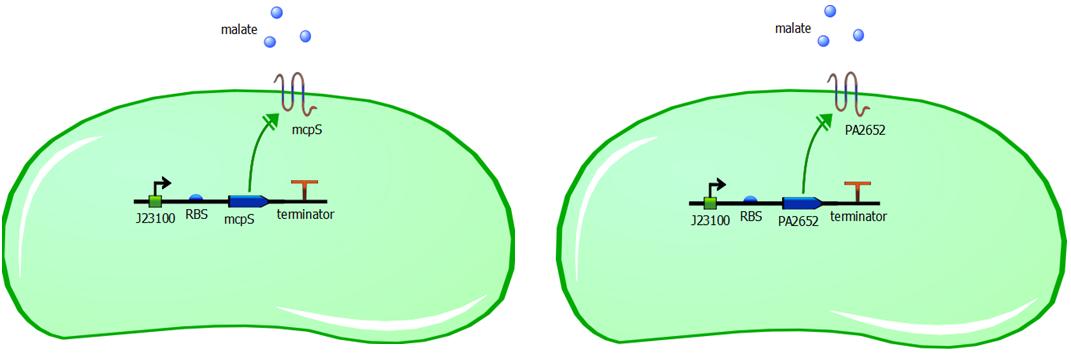
Genetic constructs for expression of malate chemoreceptor are not complicated, however they are modular. Expression of the constructs is under constitutive expression of promoter J23100. It is provided from the registry in the biobrick K398500, in the backbone vector pSB1C3. The coding sequence has been removed from this construct using PCR, providing us with backbone vector with the constitutive promoter, to be assembled using Gibson or CPEC assembly. Ribosome binding sites (RBS) have been generated using Salis RBS calculator with relative translation initiation rates (TIR): 44050 & 42010 for mcpS and PA2652 respectively. The coding sequences for mcpS and for PA2652 have been codon optimised using our codon optimisation software, so that sequence is codon optimised for E. coli and B. subtilis. However we do realise that B. subtilis is Gram-positive bacterium and therefore is not compatible with mcpS and PA2652 receptors. We exploit terminator embedded in the backbone vector pSB1C3 for termination of transcription. In addition, modularity of our system is ensured by putting 15bp insulator sequence between the RBS and the promoter. This sequence was specifically designed to make the promoter interchangeable without affecting the RBS strength and therefore translation initation rate is not affected by changing a different promoter in front of the insulator sequence. This allows us to change the promoter strength simply by changing promoter without modifying the whole genetic construct, thus it improves modularity of the construct. The insulator sequence acts as a starting point for PCR and it can be used to amplify the receptor sequence without the promoter.
Root uptake of both E. coli and the bakers’s yeast S. cerevisiae can be observed in the model organism Arabidopsis thaliana and in tomato plants. It is a process that occurs naturally (althought it yet remains to be observed in soil) and we do not need to incorporate additional genes into our design.
Modelling
Two main aspects were modelled for our chemotaxis module: malate distribution in soil and the threshold concentration of malate needed to trigger chemotaxis.
The malate concentration distribution was modelled using the Keller-Segel model.
Assembly
The receptor genes were synthesised in two fragments. In order to assemble the construct, we -use CPEC to combine the two fragments
Testing
Testing for chemotaxis can be split into qualitative and quantitative assays. Qualitative assays involve putting engineered E. coli and an attractant onto semi-solid agar plates and observe the movement of the microbes. If they can be observed to move towards the attractant source, they are likely to be attracted to the ligand. In quantitative assays, capillaries are filled with different concentrations of the attractant malate. Positive controls are provided by filling identical capillaries with different concentrations of serine, which E. coli naturally move towards. Negative controls are provided by filling capillaries with media that does not contain a source of attractant. The amount of bacteria that swim into each capillary is evaluated by FACS.
For simplicity, we will be working with Arabidopsis to observe the uptake of bacteria into plant roots. Arabidopsis thaliana is a common plant model organism. It belongs to the mustard family and fulfils many important requirements for model organisms. As such, its genome has been almost completely sequenced and replicates quickly, producing a large number of seeds. It is easily transformed and many different mutant strains have been constructed to study different aspects (National Institute of Health, no date). While Arabidopsis may not represent plant populations naturally occurring in arid areas threatened by desertification, it is a handy model organism we will be using to study the effect of auxin on roots, observe chemotaxis towards them and look at uptake of bacteria into the roots. We will be using Arabidopsis to look at the uptake of our engineered bacteria into the plants. For this, we will be using wild type Arabidopsis and E. coli that constitutively express green fluorescent protein. The natural fluorescence produced by plant roots and green fluorescence produced by the bacteria can be used to image the uptake of bacteria using confocal microscopy.
References
Chelsky, D. & Dahlquist, F. W. (1980) Chemotaxis in Escherichia coli: Association of protein components. Biochemistry, 19, 4633 – 4639.
Sourjik, V. & Armitage, J. (2010) Spatial organization in bacterial chemotaxis. The EMBO Journal, 29, 2724 - 2733.
Lacal, J., Alfonso, C., Liu, X. & et al. (2010) Identification of a chemoreceptor for tricarboxylic acid cycle intermediates: differential chemotactic response towards receptor ligands. Journal of Biological Chemistry, 285 (30), 23126 – 23136.
Alvarez-Ortega, C. & Harwood, C. S. (2007) Identification of Malate Chemoreceptor in Pseudomonas aeruginosa by Screening for Chemotaxis Defects in an Energy Taxis-Deficient Mutant. Applied and Environmental Microbiology, 73 (23, 7793 - 7795.
Paungfoo-Lonhienne et al. (2010) Turning the table: plants consume microbes as a source of nutrients. PLoS ONE 5(7): e11915.
http://www.nih.gov/science/models/arabidopsis/index.html
 "
"




