Team:Harvard/Technology/MAGE
From 2011.igem.org
(→Multiplex Automated Genome Engineering (MAGE): Editing the Bacterial Genome.) |
|||
| Line 15: | Line 15: | ||
</div> | </div> | ||
<div class="whitebox"> | <div class="whitebox"> | ||
| - | =Multiplex Automated Genome Engineering (MAGE): Editing the Bacterial Genome | + | =Multiplex Automated Genome Engineering (MAGE): Editing the Bacterial Genome= |
==How MAGE works:== | ==How MAGE works:== | ||
[[File: HARVMAGEtech1.png|right|300px]] | [[File: HARVMAGEtech1.png|right|300px]] | ||
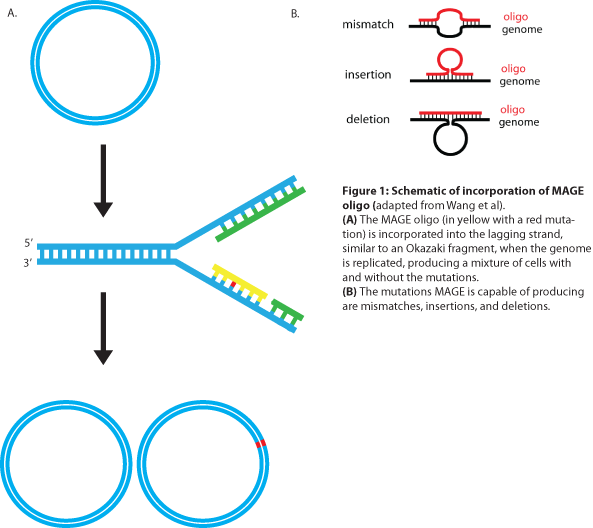
MAGE is a recently developed technique capable of editing the genome by making small changes in existing genomic sequences. This is accomplished by inserting single-stranded oligos that contain the desired mutations into the cell, and more than one gene can be targeted at a time simply by using multiple oligos. Mediated by λ-Red ssDNA-binding protein β, the oligos are incorporated into the lagging strand of the replication fork during DNA replication, creating a new allele that will spread through the population as the bacteria divide (see Figure 1). The efficiency of oligo incorporation depends on several factors, but the frequency of the allele can be increased by performing multiple rounds of MAGE on the same cell culture. Currently, researchers must perform each round by hand, a procedure requiring several hours, but automated processes are being developed to run many rounds of MAGE in succession. Together, these properties make MAGE a highly useful tool for synthetic biology, allowing researchers to easily modify the bacterial genome and generate diversity within a population. | MAGE is a recently developed technique capable of editing the genome by making small changes in existing genomic sequences. This is accomplished by inserting single-stranded oligos that contain the desired mutations into the cell, and more than one gene can be targeted at a time simply by using multiple oligos. Mediated by λ-Red ssDNA-binding protein β, the oligos are incorporated into the lagging strand of the replication fork during DNA replication, creating a new allele that will spread through the population as the bacteria divide (see Figure 1). The efficiency of oligo incorporation depends on several factors, but the frequency of the allele can be increased by performing multiple rounds of MAGE on the same cell culture. Currently, researchers must perform each round by hand, a procedure requiring several hours, but automated processes are being developed to run many rounds of MAGE in succession. Together, these properties make MAGE a highly useful tool for synthetic biology, allowing researchers to easily modify the bacterial genome and generate diversity within a population. | ||
| + | |||
| + | |||
==Oligo Design:== | ==Oligo Design:== | ||
Revision as of 14:32, 25 September 2011
MAGE | Lambda Red| Chip-Based Library | Protocols
To see how we used MAGE to build our selection strain, go to our MAGE Results section
For a step-by-step procedure, see Protocols
Contents |
Multiplex Automated Genome Engineering (MAGE): Editing the Bacterial Genome
How MAGE works:
MAGE is a recently developed technique capable of editing the genome by making small changes in existing genomic sequences. This is accomplished by inserting single-stranded oligos that contain the desired mutations into the cell, and more than one gene can be targeted at a time simply by using multiple oligos. Mediated by λ-Red ssDNA-binding protein β, the oligos are incorporated into the lagging strand of the replication fork during DNA replication, creating a new allele that will spread through the population as the bacteria divide (see Figure 1). The efficiency of oligo incorporation depends on several factors, but the frequency of the allele can be increased by performing multiple rounds of MAGE on the same cell culture. Currently, researchers must perform each round by hand, a procedure requiring several hours, but automated processes are being developed to run many rounds of MAGE in succession. Together, these properties make MAGE a highly useful tool for synthetic biology, allowing researchers to easily modify the bacterial genome and generate diversity within a population.
Oligo Design:
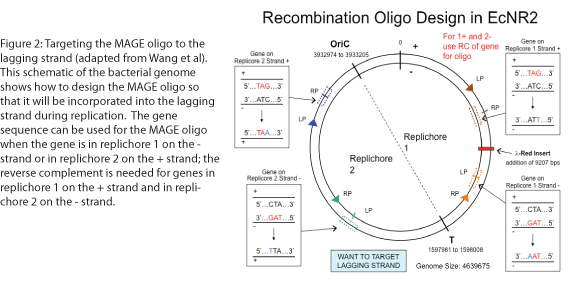
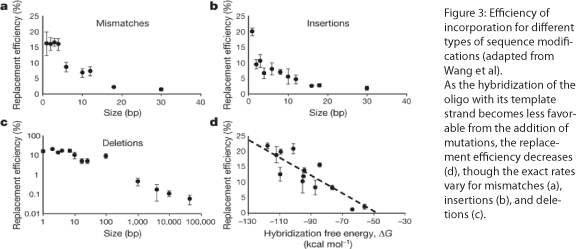
The design of the MAGE oligo is critical to the success of the procedure. Typically 90 bases long, with the first four 5’ bases phosphorothioated, the oligo must match the sequence of the region of interest (with the exception of the desired mutations) such that it will be incorporated into the lagging strand during replication. To determine which genomic strand to use as the template, it is necessary to determine whether the gene is in replichore 1 or 2 and whether it is on the + or - strand (see Figure 2). The mismatches, insertions, and/or deletions in the sequence must be centered on the oligo, and there should be as few alterations as possible, since each change will lower the efficiency of incorporation into the genome (see Figure 3).
References:
Harris H. Wang, Farren J. Isaacs, Peter A. Carr, Zachary Z. Sun, George Xu, Craig R. Forest, George M. Church. Programming cells by multiplex genome engineering and accelerated evolution. (2009). Nature, 460(7257):894-8.
 "
"