Team:Harvard/Protocols
From 2011.igem.org
(→Isothermal assembly) |
(→Protocols) |
||
| Line 5: | Line 5: | ||
__toc__ | __toc__ | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==PCR== | ==PCR== | ||
| Line 57: | Line 29: | ||
**if you are making multiple reactions, instead of adding each reagent individually to each tube, make a larger master solution of all the shared reagents in the correct proportions and then divide it among the tubes. Make sure your master solution is in excess of the amount you need (e.g. if you have 6 reactions, make a master solution with 7 or 8 times the volume for each reaction). | **if you are making multiple reactions, instead of adding each reagent individually to each tube, make a larger master solution of all the shared reagents in the correct proportions and then divide it among the tubes. Make sure your master solution is in excess of the amount you need (e.g. if you have 6 reactions, make a master solution with 7 or 8 times the volume for each reaction). | ||
**if you are doing a PCR to merely check the presence/absence of a band for a test, or to check the sizes, to conserve KAPA, you can use a total reaction volume of 10ul instead of the usual 25ul, so that one uses 5ul of KAPA per reaction, instead of 12.5ul. One can use 25-30ul when doing a PCR for PCR purification or for gel extraction | **if you are doing a PCR to merely check the presence/absence of a band for a test, or to check the sizes, to conserve KAPA, you can use a total reaction volume of 10ul instead of the usual 25ul, so that one uses 5ul of KAPA per reaction, instead of 12.5ul. One can use 25-30ul when doing a PCR for PCR purification or for gel extraction | ||
| - | |||
| - | |||
| - | |||
==Gels== | ==Gels== | ||
| Line 97: | Line 66: | ||
*Keep the orange cover on over the gel. The button at the bottom turns on a UV light so that you can see the DNA bands,and the cover protects your eyes and skin. | *Keep the orange cover on over the gel. The button at the bottom turns on a UV light so that you can see the DNA bands,and the cover protects your eyes and skin. | ||
*The UV light lets you see the gel bands, but to save a picture use the same imaging procedure as for agarose gels. | *The UV light lets you see the gel bands, but to save a picture use the same imaging procedure as for agarose gels. | ||
| - | *Be warned, however: sometimes the e-gels run inconsistently and messily... if your samples require careful discrimination between band sizes (which requires a clean gel and straight running bands), go with an agarose gel | + | *Be warned, however: sometimes the e-gels run inconsistently and messily... if your samples require careful discrimination between band sizes (which requires a clean gel and straight running bands), go with an agarose gel. |
==Cultures== | ==Cultures== | ||
| Line 141: | Line 110: | ||
*Use for miniprep, PCR purification, gel extraction | *Use for miniprep, PCR purification, gel extraction | ||
*Miniprep notes: | *Miniprep notes: | ||
| - | |||
**spin down cells (1min, 18000rcf) and remove supernatant before following kit instructions | **spin down cells (1min, 18000rcf) and remove supernatant before following kit instructions | ||
*Gel extraction notes: | *Gel extraction notes: | ||
| - | ** | + | **Stand behind plastic guard of light box and wear a disposable lab coat or long sleeves to prevent sunburn. Use a razor blade or scalpel to excise the band, but do not let any other DNA bands touch the blade (prevent contamination). Then begin the kit instructions. |
==Selection Plates== | ==Selection Plates== | ||
| Line 198: | Line 166: | ||
*Pour about 25ml into each 6cm petri dish. (Just enough to cover all of the bottom of the dish. Remember not to spill. You can invert the Bunsen burner onto the surface of the liquid agar, after pouring, to remove the air bubbles on the surface. This is so that we aren’t confused by them when distinguishing colonies from trapped air bubbles when they are used.) | *Pour about 25ml into each 6cm petri dish. (Just enough to cover all of the bottom of the dish. Remember not to spill. You can invert the Bunsen burner onto the surface of the liquid agar, after pouring, to remove the air bubbles on the surface. This is so that we aren’t confused by them when distinguishing colonies from trapped air bubbles when they are used.) | ||
*3-AT selection plates are good for only about 3 wk when stored at 4 °C. | *3-AT selection plates are good for only about 3 wk when stored at 4 °C. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==MAGE== | ==MAGE== | ||
===MAGE oligo strand choice=== | ===MAGE oligo strand choice=== | ||
| - | + | '''Choosing the strand''' | |
MAGE is much more efficient if the lagging strand is targeted with the MAGE oligo. It is thought that the MAGE oligo takes the place of the RNA primer that begins Okazaki fragments on the lagging strand during DNA replication. The diagram below shows which strand to choose when designing MAGE oligos. Use ecocyc.org[http://ecocyc.org] to determine where on the genome your site of interest lies, and which strand it's on (determined by forward or reverse orientation in the genome browser). | MAGE is much more efficient if the lagging strand is targeted with the MAGE oligo. It is thought that the MAGE oligo takes the place of the RNA primer that begins Okazaki fragments on the lagging strand during DNA replication. The diagram below shows which strand to choose when designing MAGE oligos. Use ecocyc.org[http://ecocyc.org] to determine where on the genome your site of interest lies, and which strand it's on (determined by forward or reverse orientation in the genome browser). | ||
| Line 223: | Line 175: | ||
*'''Examples''' | *'''Examples''' | ||
**For HisB deletion, we used the same strand, and for the rpoZ deletion, we used the reverse complement. | **For HisB deletion, we used the same strand, and for the rpoZ deletion, we used the reverse complement. | ||
| - | + | ||
| + | '''Protocol:''' | ||
| + | # Grow up cells to mid-log at 30C. | ||
| + | # Induce lambda red machinery by incubating cells at 42C for 15 minutes (for other strains, induction procedure may be different). | ||
| + | # Centrifuge 1-1.5mL of culture at 4C for 1 minute at top speed | ||
| + | # Remove all the media from the pellet and resuspend in 1mL of cold ddH2O before spinning again at 4C for 1 min at top speed. | ||
| + | # Repeat for a second water wash. | ||
| + | # Remove water carefully, taking care not to disturb the cell pellet. Add the MAGE oligo (amount will vary: add enough to have a final concentration of 2.5μM in a total volume of 50μL) and cold ddH2O to bring the volume up to 50μL. Mix and transfer to a cold, 1mm gap cuvette for electroporation. | ||
| + | -Note: MAGE oligo should preferentially be concentrated enough that only a few microliters need to be added. Adding larger amounts, especially if the salt concentration is high, may interfere with electroporation. | ||
| + | # Thoroughly dry the sides of the cuvette and electroporate at 1.80 kV for ideally 5.7 ms (Ec1 setting on many electroporators). | ||
| + | # Immediately add 1mL of plain LB to the cuvette, pipette up and down to mix, and transfer to a culture tube containing an additional 2mL of plain LB. | ||
| + | # Recover at 30C at least 1 hour before plating, or if performing additional rounds of MAGE, let culture grow until it reaches mid-log and repeat procedure. | ||
| + | [[File: HARVMAGEprotocol1.png]] | ||
| + | '''TAG vs TAA''' | ||
**[http://news.harvard.edu/gazette/?p=86324&utm_source=SilverpopMailing&utm_medium=email&utm_campaign=07_21_11%2520(1)&utm_content= TAG vs TAA at Church lab] | **[http://news.harvard.edu/gazette/?p=86324&utm_source=SilverpopMailing&utm_medium=email&utm_campaign=07_21_11%2520(1)&utm_content= TAG vs TAA at Church lab] | ||
| + | |||
==Isothermal assembly== | ==Isothermal assembly== | ||
Revision as of 02:19, 23 September 2011
Protocols
Contents |
PCR
temperature for annealing
[http://arep.med.harvard.edu/kzhang/cgi-bin/myOligoTm.cgi Oligonucleotide Temperature for annealing] and the Finnzymes Tm Calculator, can also be used with KAPA reactions, as the results have been better with these temperatures, with less non-specific annealing and non-specific products.
Phusion Mastermix (Finnzymes)
[http://www.neb.com/nebecomm/manualFiles/manualF-531.pdf Phusion Finnzymes]
KAPA HiFi HotStart Ready Mix (2x)
- 12.5µL KAPA readymix
- 0.75µL of each 10µM primer (final concentration 0.3µM each)
- 1ng template (for less complex DNA; for genomic DNA use 10ng)
- water up to 25µL
- initial denaturation: 95C, 2-5min
- denaturation: 98C, 20 sec
- annealing: 60-75C, 15 sec
- extension: 72C, 15 sec (for under 1kb) or 30-60 sec per kb
- cycle: 15-25 times, or if template concentration is low 30-35 cycles
- final extension: 72C, 1-5 min
- tips:
- keep master mix on ice (because of polymerase)
- if you are making multiple reactions, instead of adding each reagent individually to each tube, make a larger master solution of all the shared reagents in the correct proportions and then divide it among the tubes. Make sure your master solution is in excess of the amount you need (e.g. if you have 6 reactions, make a master solution with 7 or 8 times the volume for each reaction).
- if you are doing a PCR to merely check the presence/absence of a band for a test, or to check the sizes, to conserve KAPA, you can use a total reaction volume of 10ul instead of the usual 25ul, so that one uses 5ul of KAPA per reaction, instead of 12.5ul. One can use 25-30ul when doing a PCR for PCR purification or for gel extraction
Gels
Making a 1% agarose gel:
- Measure out 1g of agarose
- (Do not use the low melting point, LMP Agarose, since it is very fragile. Use SeaKem LE agarose)
- Pour into a flask with a lid
- Add 100mL of 1xTBE
- Microwave (use only the one on the lab bench!) for about 1.5 min with the lid loosely resting on top of the flask
- The gel will boil, but make sure it does not boil over into the microwave. You may need to stop the microwave, let the flask cool, and then continue. Keep an eye on the flash through the glass at all times. It's a good idea to put it in the microwave for 15-20s at a time, take the flask out, swirl it, and then put it back for further heating.
- The gel is done heating when it is completely clear with no white agarose flecks
- Let the gel cool until it is no longer steaming
- Add 1µL ethidium bromide (in fume hood) and swirl to mix. Caution: ethidium bromide is carcinogenic!
- Pour into a gel container with a comb and let it harden.
Loading an agarose gel:
- put gel in box and cover with 1xTBE
- Use about 10µL of ladder
- The loading buffer is 5x: add it to your sample such that the loading buffer is 1/5 the final volume
- When your sample is only a few microliters:
- stretch parafilm over an eppendorf rack
- pipet loading buffer onto parafilm (the drops will remain separate)
- pipet sample into the loading buffer drops and transfer to gel
Running agarose gels:
- typically use around 150V, but can go as high as 170V
Imaging and reading the gel for band sizes:
- use apparatus in the bay closest to the incubators. The gel should go on the UV tray, and the settings should be ethidium bromide gel, exposure for 'faint bands' if the PCR is to detect a product, and for 'intense bands' if you are loading gels for gel extraction (you would expect an intense band in that case).
- manually change exposure time if the computer-chosen one isn't adequate
- to save image: File-->export for publication-->save to the drop box with the date (e.g. 2011.06.23), a clear name, and as a png file
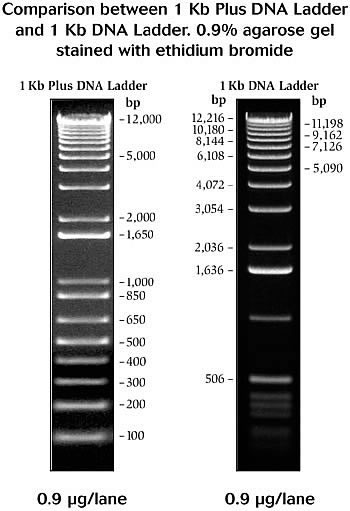
- [http://tools.invitrogen.com/content/sfs/gallery/high/2852.jpg 1 Kb Plus and 1 KB DNA ladder]
E-gels
- From experience, we find that the 1% E-gels work better than the 1.2% E-gels, and take less time (15 min instead of 20-30 min)
- E gels are much faster than agarose gels (take only 15 min) and simpler to load
- Slide gel into holder. No running buffer is needed.
- Instead of loading buffer, dilute your DNA sample up to 20µL in water. The ladder (5µL is sufficient) should also be diluted in water up to 20µL before loading.
- Keep the orange cover on over the gel. The button at the bottom turns on a UV light so that you can see the DNA bands,and the cover protects your eyes and skin.
- The UV light lets you see the gel bands, but to save a picture use the same imaging procedure as for agarose gels.
- Be warned, however: sometimes the e-gels run inconsistently and messily... if your samples require careful discrimination between band sizes (which requires a clean gel and straight running bands), go with an agarose gel.
Cultures
Liquid Culture:
- make a culture tube with 3mL LB and the appropriate antibiotic diluted to 1x (LB already with certain drugs is in iGEM fridge)
- use a pipette tip to pick up a colony from the plate and release it into the LB by stirring and pipetting up and down
- incubate at correct temperature (37 or 30C) until mid-log is reached (media slightly cloudy but not saturated, you should be able to see strings of cells swirling when you shake the tube)
- if the culture becomes saturated: you can reinoculate 30µL into a new 3mL LB tube
Glycerol stock:
- put 1200µL of a mid-log culture into a 1.5mL tube with 300µL 80% glycerol
- store at -80C (iGEM box in top row)
Smearing plates:
- pour about 10 glass beads onto the plate
- add your culture sample dropwise
- for small amounts, dilute in additional LB (100µL is plenty) so that is spreads more easily)
- shake for about 1 min so that beads spread culture around plate, changing direction frequently
- pour beads off into used beads beaker
Streaking plates:
- drop your culture on the plate
- using a pipette tip, spread the drop out into a zigzag. Then use one edge of the zigzag to draw out another zigzag. Repeat to have about 3 zigzags (this makes the culture get spread out more and more with each streak)
'Transformation:
- This process makes bacteria take up your DNA of interest. All of this (except where noted) should be done on ice, and it's a good idea to have your tubes/cuvettes sitting on ice before you use them.
- Grow your cells to mid-log: you want them to be as healthy as possible.
- If you are using lambda red, you need to induce it for 1 hr with a 0.5% total concentration of arabinose, 30˚C (time it so that after the hour the cells are still at mid-log)
- spin down culture (typically 1-1.5mL) at 4˚C
- wash pellet with 1mL of cold ddH2O twice
- resuspend with DNA and cold water up to 50µL
- electroporate: 1mm gap cuvettes (cold), 1.8kV, length of 5 units. Dry off the sides of the cuvette as well as you can before you start the current.
- immediately add 1mL plain LB to cuvette, mix, and transfer to a culture tube containing 2mL LB
- recover 2hrs in incubator
- spread on plates containing the appropriate antibiotic
[http://tools.invitrogen.com/content/sfs/manuals/oneshottop10_ecomp_man.pdf Electrocomp cells protocol]
Chemical Transformation
- Mix 1 ul of DNA to 40 ul of Top 10 One Shot ChemComp cells. Make sure to keep this on ice.
- Let the mixture sit for 30 minutes on ice.
- Heat shock the bacteria for 30 seconds at 42*C.
- Let the bacteria sit on ice for 2 minutes
- Add 480 ul of LB to the tube.
- Let the bacteria recover at 37* in the shaker for an hour.
Qiagen Kits
- Use for miniprep, PCR purification, gel extraction
- Miniprep notes:
- spin down cells (1min, 18000rcf) and remove supernatant before following kit instructions
- Gel extraction notes:
- Stand behind plastic guard of light box and wear a disposable lab coat or long sleeves to prevent sunburn. Use a razor blade or scalpel to excise the band, but do not let any other DNA bands touch the blade (prevent contamination). Then begin the kit instructions.
Selection Plates
[http://www.nature.com/nprot/journal/v1/n1/full/nprot.2006.6.html 2006, Meng, Nature Protocol]
NM medium
- 1× M9 salts
- 1x amino acid mixture containing 17 of the 20 amino acids,(store at 4°C)
- 40 mg ml–1 glucose,
- 10 µg ml–1 thiamine,
- 200 µM uracil,
- 200 µM adenine-HCl,
- 20 µM ZnSO4
- 100 µM CaCl2,
- 1 mM MgSO4.
- Filter sterilize through a 0.22-µm filter.
- NM medium can be stored at 4 °C for ~6 months
5-FOA counter-selection plates
- Autoclave 9 g Bacto agar (Difco) in 348 ml ddH2O (instead of 353ml, to make up for the changed histidine concentration) with a stir bar and then cool to 55 °C while stirring. Combine the solutions in the recipe below in order and add to the agar while stirring (final agar concentration, 1.8%, wt/vol). Use this mixture (500 ml final volume) to pour five 245×245–mm square plates (100 ml per plate).
- 20× M9 salt solution 25 ml
- 1% (wt/vol) yeast extract 5 ml (store at 4°C)
- 40% (wt/vol) glucose 5 ml
- 20 mM uracil 5 ml
- 5% (wt/vol) histidine 10 ml (instead of 5 ml of 10% (wt/vol) histidine solution) (keep at room temperature, this tends to crystalise out of solution)
- 0.1 M CaCl2 0.5 ml
- 1 M MgSO4 0.5 ml
- 1% (wt/vol) thiamine 0.5 ml
- 20 mM ZnSO4 0.25 ml
- 12.5 mM 5-FOA 100 ml (TO DO)
- 50 mg ml–1 kanamycin 0.25 ml
- The 5-FOA plates can be stored at 4 °C for a few weeks.
3-AT selection plates
- Autoclave 9 g bacto agar in 443 ml ddH2O with a stir bar. Stir agar while cooling to ~55 °C.
- Combine the solutions in the recipe below in order and add to the agar.
- 20× M9 salt solution - 25 ml
- 40% (wt/vol) glucose - 5 ml
- 20 mM adenine - 5 ml
- 33.3× amino acid mixture - 15 ml
- 20 mM uracil - 5 ml
- 0.1 M CaCl2 - 0.5 ml
- 1 M MgSO4 - 0.5 ml
- 1% (wt/vol) thiamine - 0.5 ml
- 20 mM ZnSO4 - 0.25 ml
- Then add
- 50 µl 100 mM IPTG (10 µM final concentration, our stock solution of 500mM IPTG was first diluted 5x to 100mM concentration),
- 0.5 to 2.5 ml of 1 M 3-AT (depending on the final desired concentration of 3-AT. N.B. We used two concentrations here, one low with 0.5ml, and one in the middle with 1.5ml, respectively),
- 0.25 ml of 50 mg ml–1 kanamycin (N.B. We used 2.5ml of 5 mg ml–1 kanamycin, which is our 1000x stock solution) for a dilution to 25 µg ml–1 final concentration and
- 0.5 ml 10 mg ml–1 spectinomycin
- to the agar while stirring (500 ml final volume).
(If the pB1H2 plasmid is also being used, add carbenicillin to a final concentration of 100 µg ml–1. N.B. This is not applicable as we don’t use this plasmid, so don’t add carbenicillin/ampicillin.)
- Pour about 25ml into each 6cm petri dish. (Just enough to cover all of the bottom of the dish. Remember not to spill. You can invert the Bunsen burner onto the surface of the liquid agar, after pouring, to remove the air bubbles on the surface. This is so that we aren’t confused by them when distinguishing colonies from trapped air bubbles when they are used.)
- 3-AT selection plates are good for only about 3 wk when stored at 4 °C.
MAGE
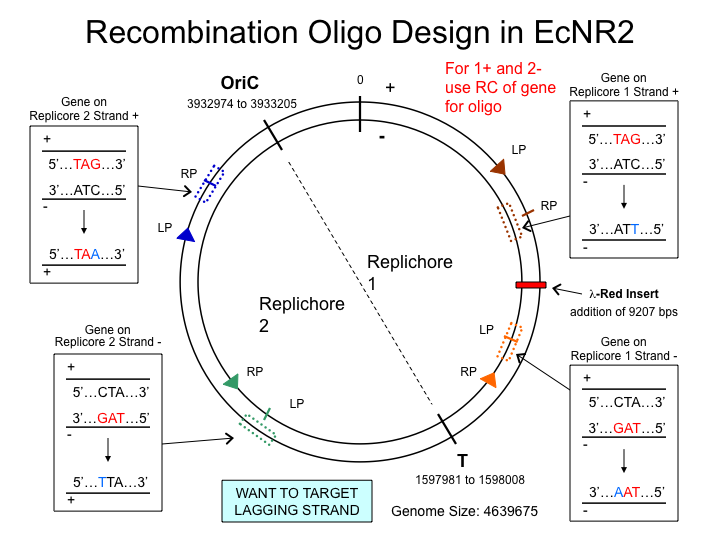
MAGE oligo strand choice
Choosing the strand
MAGE is much more efficient if the lagging strand is targeted with the MAGE oligo. It is thought that the MAGE oligo takes the place of the RNA primer that begins Okazaki fragments on the lagging strand during DNA replication. The diagram below shows which strand to choose when designing MAGE oligos. Use ecocyc.org[http://ecocyc.org] to determine where on the genome your site of interest lies, and which strand it's on (determined by forward or reverse orientation in the genome browser).
- Examples
- For HisB deletion, we used the same strand, and for the rpoZ deletion, we used the reverse complement.
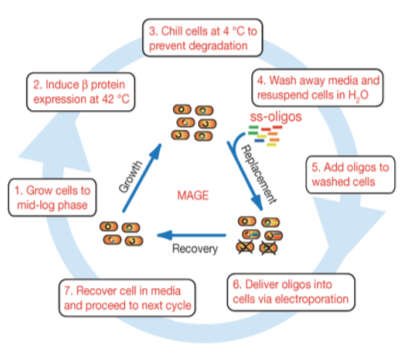
Protocol:
- Grow up cells to mid-log at 30C.
- Induce lambda red machinery by incubating cells at 42C for 15 minutes (for other strains, induction procedure may be different).
- Centrifuge 1-1.5mL of culture at 4C for 1 minute at top speed
- Remove all the media from the pellet and resuspend in 1mL of cold ddH2O before spinning again at 4C for 1 min at top speed.
- Repeat for a second water wash.
- Remove water carefully, taking care not to disturb the cell pellet. Add the MAGE oligo (amount will vary: add enough to have a final concentration of 2.5μM in a total volume of 50μL) and cold ddH2O to bring the volume up to 50μL. Mix and transfer to a cold, 1mm gap cuvette for electroporation.
-Note: MAGE oligo should preferentially be concentrated enough that only a few microliters need to be added. Adding larger amounts, especially if the salt concentration is high, may interfere with electroporation.
- Thoroughly dry the sides of the cuvette and electroporate at 1.80 kV for ideally 5.7 ms (Ec1 setting on many electroporators).
- Immediately add 1mL of plain LB to the cuvette, pipette up and down to mix, and transfer to a culture tube containing an additional 2mL of plain LB.
- Recover at 30C at least 1 hour before plating, or if performing additional rounds of MAGE, let culture grow until it reaches mid-log and repeat procedure.
- [http://news.harvard.edu/gazette/?p=86324&utm_source=SilverpopMailing&utm_medium=email&utm_campaign=07_21_11%2520(1)&utm_content= TAG vs TAA at Church lab]
Isothermal assembly
Courtesy of Megason lab Wiki (Thanks to Nikolaus Obholzer - 22 Sep 2009)
Isothermal Assembly
Isothermal Assembly works by combining a cocktail of exonuclease, polymerase, and ligase to fuse dsDNA fragments with sufficiently (20-120 bp) homologous ends. It leaves no "scar" behind, i.e. you can expect your product to contain the EXACT overlap sequence. The reaction may work with shorter ends (e.g. 15 bp), so long as the annealing temperature is higher than 50C.
Isothermal assembly reactions are stored in the common -20C freezer as 7.5 ul aliquots (in PCR strip tubes).
To perform isothermal assembly:
1. PCR up your fragments of choice, and gel purify. High concentration DNA is critical, since it needs to fit in a small volume!
2. Not exceeding a total volume of 2.5 ul (can be a bit more, maybe up to 3.5 ul), in a PCR tube, combine fragments at equal molecular ratio [e.g. amount fragment1 = 100 ng * (fragment size 1/ fragment size 2); amount fragment2 = 100 ng * (fragment size 2/ fragment size 1) ... etc.]. If required, bring to 2.5 ul with ddH2O. I recommend using approx. 100 ng of plasmid backbone (fragment containing antibiotic resistance).
3. Add the combined fragments (5 ul) [2.5 ul] to 1 Isothermal Assembly reaction aliquot (15 ul) [We use 7.5 ul instead] and mix by pipetting (20 ul total) [10 ul total].
4. Place rxn. at 50C for 15 -60 min [We put it in a thermocycler for 60 min @ 50C, then 4C forever].
5. (optional for chem. transform.) Purify with Qiagen PCR purification (MinElute) kit. Elute in 20 ul of ddH2O.
6. Transform with 1 ul of assembly rxn.
References: Gibson et al (2009) Nature Methods 6(5):343-345.
Gibson_and_Smith_2009_-_Enzymatic_assembly_of_DNA_molecules_up_to_several_hundred_kilobases.pdf: Reference paper for isothermal assembly
Making isothermal assembly aliquots
5x isothermal assembly reaction buffer (assemble on ice):
From the paper: Actually added:
- 3 mL 1M Tris-HCl pH 7.5 3 mL 1M Tris-HCl pH 7.5
- 150 uL 2M MgCl2 300 uL 1M MgCl2
- 60 uL 100 mM dGTP 600 uL 10 mM each dNTP
- 60 uL 100 mM dCTP
- 60 uL 100 mM dTTP
- 60 uL 100 mM dATP
- 300 uL 1M DTT 300 uL 1M DTT
- 1.5 g PEG-8000 1.5 g PEG-8000
- 300 uL 100 mM NAD 20 mg NAD
- ddH2O to 6 mL ddH2O to 6 mL
Prepare 320 uL aliquots (18) and freeze all but one. Label these “5X isotherm buffer”
To the one remaining (320 uL), add:
- 1.2 uL T5 Exonuclease
- 20 uL Phusion polymerase (NOT HOTSTART)
- 160 uL Taq ligase
- 700 uL ddH2O
Prepare 7.5 uL aliquots (~160) on ice in PCR tubes and store at -20C. These should be good for up to a year.
Gel extraction
Gel extraction allows you to concentrate a desired PCR product. It is required for isothermal assembly. Use [http://www.qiagen.com/literature/render.aspx?id=537 the protocol provided with the Qiagen kit] with the following changes:
Step Change
2. Add only 2 gel volumes of buffer QG.
3. May need to incubate longer at 50*C (for approximately 15 to 20 minutes)
4. Add 10 ul for 3M NaOAc. This will correct the pH and should turn the QG back to yellow.
7. Combine samples of identical DNA in the same column (spin multiple times if necessary).
9. Skip this step.
13. Elute in 30 ul rather than 50 ul. Use molecular grade H2O instead of Buffer ED to really maximize yield (not a necessary protocol change,
though). Let column stand for 1 to 2 minutes before spinning.
PCR purification
Use [http://www.qiagen.com/literature/render.aspx?id=201082 the protocol] provided with the Qiagen kit with the following changes:
Step Change
1 Mix all of the PCR product in 500 ul of Buffer PB.
7 Elute in 30 ul rather than 50 ul. Use molecular grade H2O instead of Buffer EB to really maximize yield (not a necessary protocol change,
though). Let column stand for 1 to 2 minutes before spinning.
Lambda Red
Lambda Red [http://openwetware.org/wiki/Recombineering/Lambda_red-mediated_gene_replacement]
Lambda Red Recombination
- Activate lambda red through temperature sensitive activation
- Add 1-1.5 mL of mid-log cells in culture to 1.5 mL eppendorf tube
- All steps need to be done on ice and must be chilled at all times!!
- Wash cells with 1 mL of cold H2O, and spin in centrifuge for 1 minute at max speed
- Wash cells twice
- While removing water after spinning down, helps to hold tube so bead is at the top and then suck out water below it
- Resuspend cells with 50 μL of cold H2O
- Add 50-200 ng of insertion construct to eppendorf
- Then place in chilled cuvette
- Set electroporation machine to Eco1, 1.8 volts, and time constant around 5 ms for good electroporation
- Before make sure that all water is whiped off of cuvette, or else polar water molecules can interfere
- After electroporation immediately place 1 mL of LB in cuvette
- Then place 1 mL of LB and product into a 2 mL of LB and allow to grow for 2 to 3 hours in correct temperature
- After three hours, plate out culture in different concentrations on specific plates
Chip DNA Extraction
- See [http://www.nature.com/nbt/journal/v28/n12/extref/nbt.1716-S1.pdf Supplementary material] from Kosuri et al 2010
Shopping List
Before you buy, first make sure we actually have run out of these reagents. Here are some suggestions for where to procure suitable reagents:
Buffer
Item Vendor Cat. No.
- 1M Tris-HCl pH 7.5 Make in-house none
- Magnesium Chloride, 1.00 +/- 0.01M Solution Affymetrix / USB 78641 10 x 1 ML
- Nicotinamide adenine dinucleotide (NAD) Applichem A1124,0005
- DTT, molecular biology grade FERMENTAS R0861
- Polyethylene Glycol 8000, Powder USB / Affymetrix 19966
- dNTP Mix, 10mM each Fermentas R0192 1 ml
Enzymes:
- T5 Exonuclease EPICENTRE T5E4111K
- Taq DNA Ligase NEB M0208L
- Phusion™ High-Fidelity DNA Polymerase NEB F-530S
References: Gibson et al (2009) Nature Methods 6(5):343-345.
 "
"