Team:Groningen/modeling simulation engine
From 2011.igem.org
| Line 22: | Line 22: | ||
[[File:TeamGroningenSimulatorUML.png|centre|700px|A simple UML diagram of the different modeling component.]] | [[File:TeamGroningenSimulatorUML.png|centre|700px|A simple UML diagram of the different modeling component.]] | ||
| - | |||
| - | |||
| - | |||
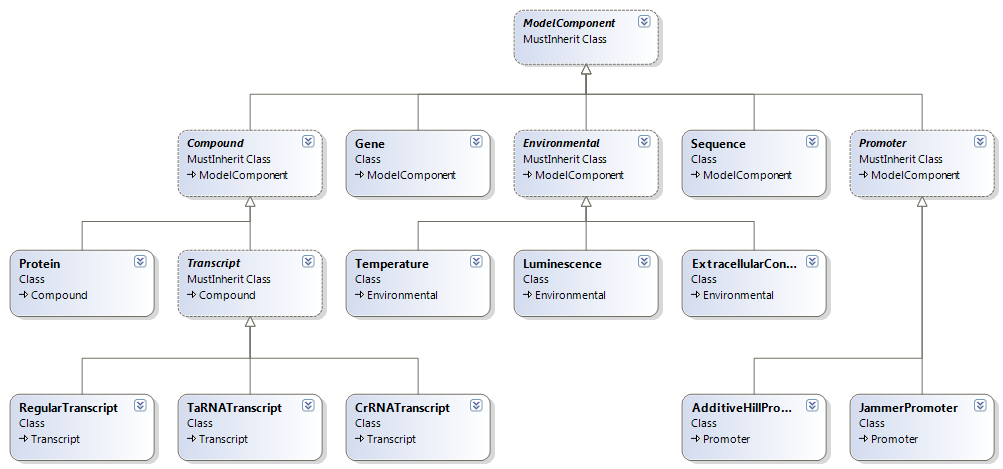
| - | + | *ModelComponent: The abstract base class of all model components. All ModelComponents have a name. | |
| - | + | ||
| - | + | *Compound: The abstract base class of all the stuff floating around inside the cell. Compounts all have a concentration which is registered as nano moles for every timestep. | |
| - | + | ||
| - | + | ||
| - | + | *Protein: The class we used for everything that is a compound but not a transcript. Proteins have a concentration decay rate over time, which covers degredation as well as the dilution because of cell growth. We used this class not only for proteins but also for smaller molecules that we assumed shared this behavoir (such as arabinose or IPTG). | |
| + | *Transcript: The abstract base class for all transcripts. We treat everything that is a direct result of a transcription process. Every Transcript has a concentration decay rate. It has several subfunctionsclasses that express the different types of transcript we modeled | ||
| - | + | ||
| - | + | *Regular Transcript: This is a transcript that moves on to translation. It has a Ribosome binding site | |
| - | + | *taRNA Transcript | |
| + | *crRNA Transcript | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
==Model== | ==Model== | ||
| Line 53: | Line 44: | ||
* Transcript | * Transcript | ||
| - | * | + | * |
| - | + | ||
| - | + | ||
* Promoter | * Promoter | ||
Revision as of 09:41, 20 September 2011
The Cumulus simulation engine
Cumulus uses an internal single cell simulation engine to evaluate parameter settings. The simulation engine is a simple temporal integration over a collection of differential equations. As an input it takes a model of the cell to simulate, some starting conditions, and the input data relating to controlled environmental factors. The timescale we use is seconds since this seems a good middle ground that allows us to characterize the processes we are interested in typically the effects of reporters such as GFP. All faster processes such as diffusion of compound through the cell or the binding of induction factors to operating regions are assumed to occur instantaneously between time steps. Slower processes such as transcription and translation delays are explicitly modeled. The reason we do not allow the use of variable timescales is a these can yield numerically different solutions for the same data. While try to remain consistend between models but within the context of our modeling tool.
The model
For our model we deterministic set of differential equations (ODE's and PDE's). All these act on the current model state to compute the next state of the model. This state represents all the important chacarteristics in a cell which we refer to as factors.
We distingish four types of factors:
- Promoter activations, measures in polimerase bindings per second, for instance the activation of a LacI promoter
- Transcript concentrations, measured in nanomole per liter, for instance the concentration of an RNA molecule coding for cI.
- Compound concentrations, measured in nanomole per liter, for instance the concentration of an GFP molecule.
- Environmental factors, measured in different units, for instance temprature or ligth exposure.
Using these four types of factors we can generate new models states. By saving every model state we can plot the behavior of our system in a esealy readable way if needed.
[Illustration]
Modularety
The Cumulus simulation engine is a system is of modular object oriened design. Because of this adding new components can be done in a matter of minutes. We only implemented the components we needed for our own system and and some test construcs, but the potential for extendabilety is very large. At the moment the simulators biggest contraines are a lack of noise modeling and multy cellular support.
- ModelComponent: The abstract base class of all model components. All ModelComponents have a name.
- Compound: The abstract base class of all the stuff floating around inside the cell. Compounts all have a concentration which is registered as nano moles for every timestep.
- Protein: The class we used for everything that is a compound but not a transcript. Proteins have a concentration decay rate over time, which covers degredation as well as the dilution because of cell growth. We used this class not only for proteins but also for smaller molecules that we assumed shared this behavoir (such as arabinose or IPTG).
- Transcript: The abstract base class for all transcripts. We treat everything that is a direct result of a transcription process. Every Transcript has a concentration decay rate. It has several subfunctionsclasses that express the different types of transcript we modeled
- Regular Transcript: This is a transcript that moves on to translation. It has a Ribosome binding site
- taRNA Transcript
- crRNA Transcript
Model
=Basic types
- Sequence
- Compound
- Transcript
- Promoter
- Additive Hill Promoter
- Jammed Promoter
- Gene
- Protein
- Temperature
- Extracellular Concentration
 "
"