Team:Groningen/project degradation tags
From 2011.igem.org
Degradation Tags
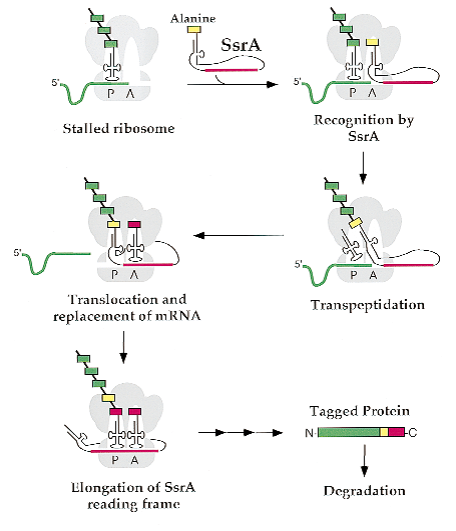
Bacteria are able to synthesize a special RNA called SsrA RNA which is able to function both as tRNA and mRNA during the protein synthesis. This RNA fragment acts when there is an incomplete or untranslatable part in the RNA being translated. Translation stalls which ends up with SsrA (which has a charged alanine group that binds to the polypeptide chain leaving the ribosome) being recognized by the ribosome as a tRNA. SsrA binds to the ribosome and the message is changed to the tRNA part of SsrA which codes for a degradation tail, in our case an LVA tag. An LVA tag has an amino acid sequence of AANDENYALVA. In bacteria, this tail acts as a precursor for fast degradation. Some chaperons found in the same bacteria, mainly ClpAP and ClpXP, recognize this tail and degrade this wrongly translated polypeptide. In this way, the bacterium can get rid of the unwanted product which may even harm itself. In our project, we did not use SsrA to add the degradation tag, but we synthetically added the degradation tag to the DNA sequence of the proteins that are needed to be expressed for a short amount of time. ClpXP is able to recognize these proteins and degrade them in a short amount of time.
 "
"