Team:Groningen/modeling results
From 2011.igem.org
Results
Fitting
In its most simple use the Cumulus system can be used to characterize promoters. The example below shows how this can be done for the Pbad Promoter with flowcytometry data. The measurements points in this case are generated by comparing as measurement of an unknown part against two measurements from a well characterized one.
movie
Dim Arabinose As Compound = New Protein(ParameterSetting, "Arabinose") Dim GFP As Compound = New Protein(ParameterSetting, "GFP")
Dim GFPGene As Gene = New Gene(ParameterSetting, "GFPGene", GFP) Dim PbadPromoter As AdditiveHillPromoter = New AdditiveHillPromoter(ParameterSetting, "Pbad Promoter", "Arabinose")
Dim GFPTranscript As Transcript = New RegularTranscript(ParameterSetting, "GFP Transcript", GFPGene) Dim PbadSequence As Sequence = New Sequence(ParameterSetting, "Pbad Sequence", PbadPromoter, GFPTranscript) Dim Sequences As List(Of Sequence) = New List(Of Sequence) Sequences.Add(PbadSequence)
Dim Compounds As List(Of Compound) = New List(Of Compound) Compounds.Add(Arabinose) Compounds.Add(GFP)
Dim Environmentals As List(Of Environmental) = New List(Of Environmental) Environmentals.Add(New ExtracellularConcentration(ParameterSetting, "Arabinose"))
' Output Dim SensorBase As Double = 0 Dim SensorMultiplier As Double = 500 Environmentals.Add(New Luminescence(ParameterSetting, "Green Fluorescence", "GFP", SensorBase, SensorMultiplier))
Dim Cell As Cell = New Cell(Sequences, Compounds, Environmentals) Return New Model(Cell)
Public Overrides Function GetInitialParameters() As ParameterSetting
Dim ParameterSetting As ParameterSetting = New ParameterSetting()
ParameterSetting.Add("Pbad Promoter", "LeakageActivation", 0.0001, 0, 0.01)
ParameterSetting.Add("Pbad Promoter", "MaximumActivation", 0.02, 0.01, 0.02)
ParameterSetting.Add("Pbad Promoter", "KMid", 20000, 10000, 40000)
ParameterSetting.Add("Pbad Promoter", "Cooperativety", 2, 1, 3)
ParameterSetting.Add("Arabinose", "Halflife", 2520, 600, 3600)
ParameterSetting.Add("GFP", "Halflife", 2520, 600, 3600)
ParameterSetting.Add("GFP Transcript", "Halflife", 240, 180, 300)
ParameterSetting.Add("GFP Transcript", "RIBS", 0.02, 0, 0.1)
ParameterSetting.Add("GFP Transcript", "Delay", 300, 120, 600)
ParameterSetting.Add("Pbad Sequence", "CopyNumber", 10, 9, 11)
ParameterSetting.Add("Pbad Sequence", "TranscriptionTime", 300, 120, 600)
ParameterSetting.Add("Extracellular Arabinose", "TransportRate", 0.00001, 0, 1)
Return ParameterSetting
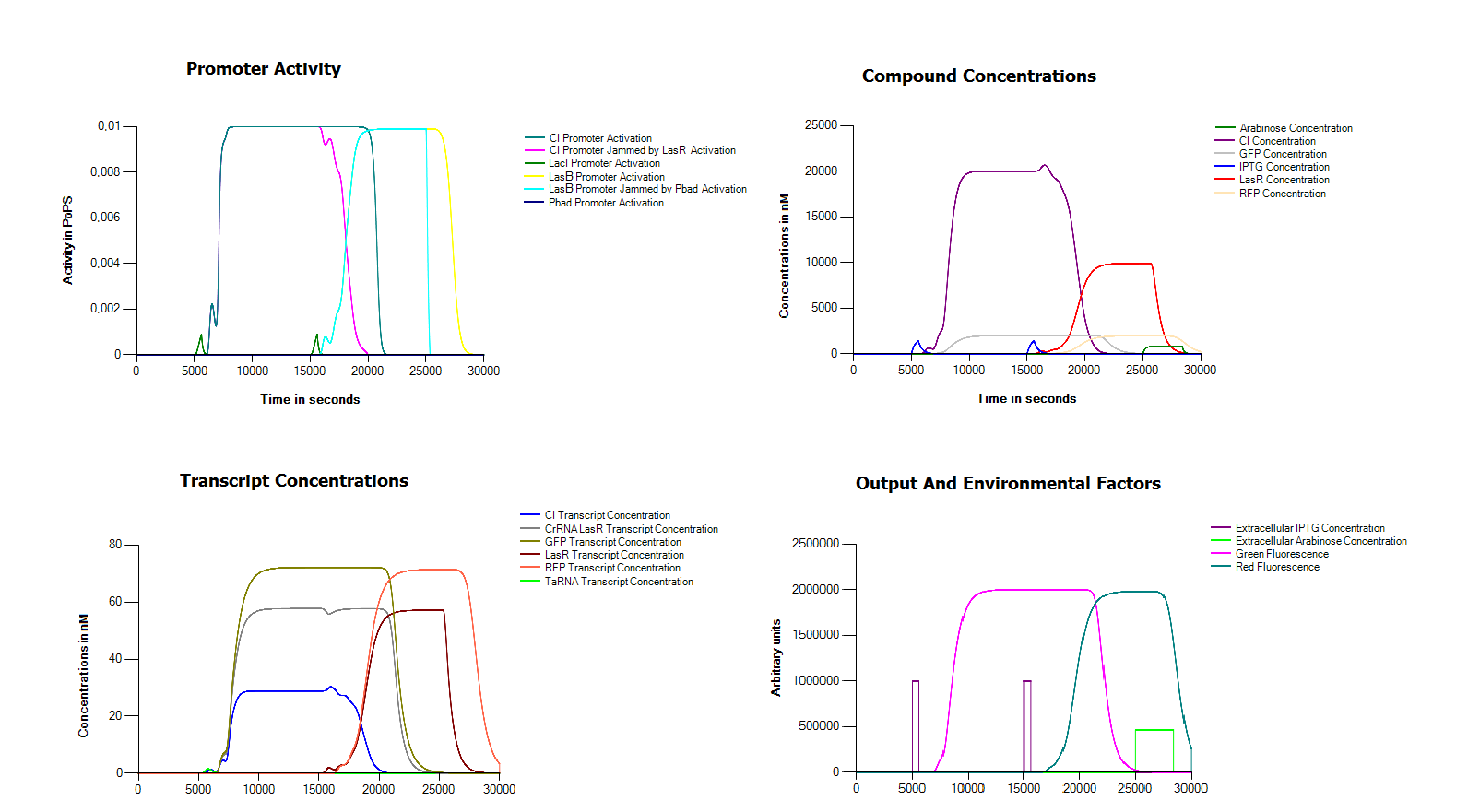
Visulisation of out model
Because we did not manage to finish our model we decided to try to model our system in the Cumulus system. One of the main problems in assembling our system seemed to be the defective hybB promoter. For this reason in this model we replaced it with a LacI promoter. In order to operate our system we also assume that we can remove both arabinose and IPTG from the medium in an effcient way.
Speedup
The Cumulus system allows for large speedups by parallelization through both the cloud and added machines. We tried this with some typical cloud intances and some local machines in the lab, the results are listed below. The benchmarking was done the the Pbad characterization task described above.
 "
"