Team:EPF-Lausanne/Protocols/TetR Extension PCR
From 2011.igem.org
(→Mutation PCR) |
|||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:EPF-Lausanne/Templates/ | + | {{:Team:EPF-Lausanne/Templates/ProtocolHeader|title=TetR Extension PCR}} |
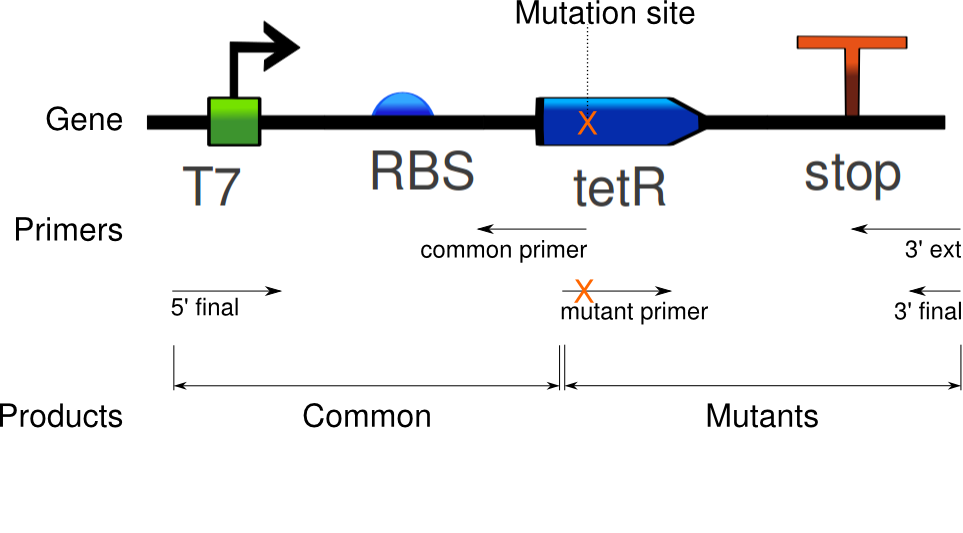
The purpose of this procedure is to induce specific mutations in the tetR linear template. It requires two sets of PCRs: the first set amplifies the ''common sequence'' and the ''mutant sequence'', and induces mutations in the mutant sequence. As illustrated below, the common sequence extends from the beginning of the gene to the site of mutations. The mutation sequence covers the remaining segment. The second set of PCRs stitches the two sequences together, making a mutant gene. In the following, the first set of PCRs is referred to as "Mutation PCR", and the second set as "Fusion PCR". | The purpose of this procedure is to induce specific mutations in the tetR linear template. It requires two sets of PCRs: the first set amplifies the ''common sequence'' and the ''mutant sequence'', and induces mutations in the mutant sequence. As illustrated below, the common sequence extends from the beginning of the gene to the site of mutations. The mutation sequence covers the remaining segment. The second set of PCRs stitches the two sequences together, making a mutant gene. In the following, the first set of PCRs is referred to as "Mutation PCR", and the second set as "Fusion PCR". | ||
| Line 7: | Line 7: | ||
In the '''mutation PCR''', one reaction has to be run for the common sequence, then one reaction per mutant. For example, seven reactions are needed for six mutants. All of them use the tetR linear template; they differ by their primers. The common sequence uses primers ''5' final'' and ''common primer'', whereas the mutant sequences use primers ''3' ext'' and ''mutant primer''. All but the mutant primer are standard PCR primers (XXX with overlap? XXX). The mutant primer is the key to inducing mutations: it doesn't perfectly match the sequence to copy, but contains one or more substitutions that are then replicated on the PCR product. After this PCR, the desired sequences are gel extracted, and used for the next step. | In the '''mutation PCR''', one reaction has to be run for the common sequence, then one reaction per mutant. For example, seven reactions are needed for six mutants. All of them use the tetR linear template; they differ by their primers. The common sequence uses primers ''5' final'' and ''common primer'', whereas the mutant sequences use primers ''3' ext'' and ''mutant primer''. All but the mutant primer are standard PCR primers (XXX with overlap? XXX). The mutant primer is the key to inducing mutations: it doesn't perfectly match the sequence to copy, but contains one or more substitutions that are then replicated on the PCR product. After this PCR, the desired sequences are gel extracted, and used for the next step. | ||
| - | The '''fusion PCR''' is then run with the common sequence and one | + | The '''fusion PCR''' is then run with the common sequence and one mutant sequences as templates, and primers ''5' final'' and ''3' final''. Each mutant is reacted separately. |
== Equipment needed == | == Equipment needed == | ||
| Line 23: | Line 23: | ||
== Mutation PCR == | == Mutation PCR == | ||
| - | One reaction is run per mutant, plus one for the common sequence, with the reagents listed below. Mix in individual PCR tubes. The ''mutant primer'' is different for every mutant sequence. | + | * One reaction is run per mutant, plus one for the common sequence, with the reagents listed below. Mix in individual PCR tubes. The ''mutant primer'' is different for every mutant sequence. |
'''Warning''': mix the polymerase last, and make sure the thermal cycler is available before mixing it. | '''Warning''': mix the polymerase last, and make sure the thermal cycler is available before mixing it. | ||
| Line 69: | Line 69: | ||
|} | |} | ||
| - | Run the PCR with the following thermal cycles: | + | * '''Run the PCR''' with the following thermal cycles: |
{| | {| | ||
| Line 90: | Line 90: | ||
|} | |} | ||
| - | Upon completion of the PCR, run a [[Team:EPF-Lausanne/Protocols/Gel_Electrophoresis|gel electrophoresis]] with 5 ul of PCR product per well, using 1 kb and 100 kb ladder. If the results are satisfactory, extract the products by [[Team:EPF-Lausanne/Protocols/Gel_purification|gel extraction]]. | + | * Upon completion of the PCR, run a [[Team:EPF-Lausanne/Protocols/Gel_Electrophoresis|gel electrophoresis]] with 5 ul of PCR product per well, using 1 kb and 100 kb ladder. |
| + | * If the results are satisfactory, extract the products by [[Team:EPF-Lausanne/Protocols/Gel_purification|gel extraction]]. | ||
| + | * The end result should be a set of mutant sequences and a common sequence, dissolved EB buffer, in separate tubes. | ||
== Fusion PCR == | == Fusion PCR == | ||
| + | {| | ||
| + | |+ Reagents for the fusion PCR | ||
| + | ! Reagent || Vol. [ul] | ||
| + | |- | ||
| + | | Primer 5' final (250 uM) || 1 | ||
| + | |- | ||
| + | | Primer 3' final (250 uM) || 1 | ||
| + | |- | ||
| + | | Product of common seq. PCR || 0.5 | ||
| + | |- | ||
| + | | Product of mutant seq. PCR || 0.5 | ||
| + | |- | ||
| + | | dNTP || 1 | ||
| + | |- | ||
| + | | MF buffer iProof 5x || 10 | ||
| + | |- | ||
| + | | iProof polymerase || 0.5 | ||
| + | |- | ||
| + | | dH20 || 35.5 | ||
| + | |- | ||
| + | | total volume: || 50 | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | {| | ||
| + | |+ Thermal cycles for the fusion PCR | ||
| + | ! # || T [°C] || Duration | ||
| + | |- | ||
| + | | 1 || 98 || 30 s | ||
| + | |- | ||
| + | | 2 || 98 || 8 s | ||
| + | |- | ||
| + | | 3 || 55 || 20 s | ||
| + | |- | ||
| + | | 4 || 72 || 15 s | ||
| + | |- | ||
| + | | || Repeat 2-4 30x || | ||
| + | |- | ||
| + | | 5 || 72 || 5 min. | ||
| + | |- | ||
| + | | 6 || 10 || forever | ||
| + | |} | ||
| - | {{:Team:EPF-Lausanne/Templates/ | + | {{:Team:EPF-Lausanne/Templates/Footer}} |
Latest revision as of 08:15, 20 July 2011
TetR Extension PCR
Back to protocols.The purpose of this procedure is to induce specific mutations in the tetR linear template. It requires two sets of PCRs: the first set amplifies the common sequence and the mutant sequence, and induces mutations in the mutant sequence. As illustrated below, the common sequence extends from the beginning of the gene to the site of mutations. The mutation sequence covers the remaining segment. The second set of PCRs stitches the two sequences together, making a mutant gene. In the following, the first set of PCRs is referred to as "Mutation PCR", and the second set as "Fusion PCR".
In the mutation PCR, one reaction has to be run for the common sequence, then one reaction per mutant. For example, seven reactions are needed for six mutants. All of them use the tetR linear template; they differ by their primers. The common sequence uses primers 5' final and common primer, whereas the mutant sequences use primers 3' ext and mutant primer. All but the mutant primer are standard PCR primers (XXX with overlap? XXX). The mutant primer is the key to inducing mutations: it doesn't perfectly match the sequence to copy, but contains one or more substitutions that are then replicated on the PCR product. After this PCR, the desired sequences are gel extracted, and used for the next step.
The fusion PCR is then run with the common sequence and one mutant sequences as templates, and primers 5' final and 3' final. Each mutant is reacted separately.
Contents |
Equipment needed
A standard PCR setup is used.
Other techniques necessary:
Primer Design
TODO. Get primers, find out how they were designed. Considerations, constraints, and tools.
Mutation PCR
- One reaction is run per mutant, plus one for the common sequence, with the reagents listed below. Mix in individual PCR tubes. The mutant primer is different for every mutant sequence.
Warning: mix the polymerase last, and make sure the thermal cycler is available before mixing it.
| Reagent | Vol. [ul] |
|---|---|
| Primer 5' final (100 uM) | 0.5 |
| Primer common (100 uM) | 0.5 |
| tetR linear template (100x diluted) | 0.5 |
| dNTP | 1 |
| MF buffer iProof 5x | 10 |
| iProof polymerase | 0.5 |
| dH20 | 37 |
| total volume | 50 |
| Reagent | Vol. [ul] |
|---|---|
| Primer 3' ext (100 uM) | 0.5 |
| Mutant primer (100 uM) | 0.5 |
| tetR linear template (100x diluted) | 0.5 |
| dNTP | 1 |
| MF buffer iProof 5x | 10 |
| iProof polymerase | 0.5 |
| dH20 | 37 |
| total volume | 50 |
- Run the PCR with the following thermal cycles:
| # | T [°C] | Duration |
|---|---|---|
| 1 | 98 | 30 s |
| 2 | 98 | 7.5 s |
| 3 | 58 | 20 s |
| 4 | 72 | 15 s |
| Repeat 2-4 30x | ||
| 5 | 72 | 5 min. |
| 6 | 10 | forever |
- Upon completion of the PCR, run a gel electrophoresis with 5 ul of PCR product per well, using 1 kb and 100 kb ladder.
- If the results are satisfactory, extract the products by gel extraction.
- The end result should be a set of mutant sequences and a common sequence, dissolved EB buffer, in separate tubes.
Fusion PCR
| Reagent | Vol. [ul] |
|---|---|
| Primer 5' final (250 uM) | 1 |
| Primer 3' final (250 uM) | 1 |
| Product of common seq. PCR | 0.5 |
| Product of mutant seq. PCR | 0.5 |
| dNTP | 1 |
| MF buffer iProof 5x | 10 |
| iProof polymerase | 0.5 |
| dH20 | 35.5 |
| total volume: | 50 |
| # | T [°C] | Duration |
|---|---|---|
| 1 | 98 | 30 s |
| 2 | 98 | 8 s |
| 3 | 55 | 20 s |
| 4 | 72 | 15 s |
| Repeat 2-4 30x | ||
| 5 | 72 | 5 min. |
| 6 | 10 | forever |
 "
"