Team:EPF-Lausanne/Notebook/September2011
From 2011.igem.org
Notebook: September 2011
Thursday, September 1 2011
Vincent PCR purified the pSB3K1-extended (no TetR) for use in the Gibsons and negative controls.
The Gibson assembly of J6-Plac-Lysis had produced a single colony. Vincent colony-PCRed it using two different sets of primers to amplify subportions of the lysis cassette. The first set of primers (30-f & 816-r) amplify an 800 bp piece whereas the second set (Seq-J6-RFP-f & Seq-J6-RFPlysis-r) amplifies a 1100 bp piece.
The gel seems to confirm that lysis is in the plasmid. We will send the plasmid for sequencing on Friday.
After taking a closer look at the plate from the platereader, it seems that none of the T7 variants did any lysing. This realization made it urgent that we discover whether or not the full T4-lysis cassette was really in these Gibson-assembled plasmids. The samples that had been sent in for sequencing had primers that seemed to amplify the backbone, instead of the lysis cassette. While this does not necessarily mean that the lysis is not there, it does suggest that all the PCRS and gels used to verify the presence of lysis were only showing the existence of the backbone.
Alina ran a PCR with different primers from Douglas' first attempts at putting together the 2700 bp cassette that amplify select regions of the cassette.
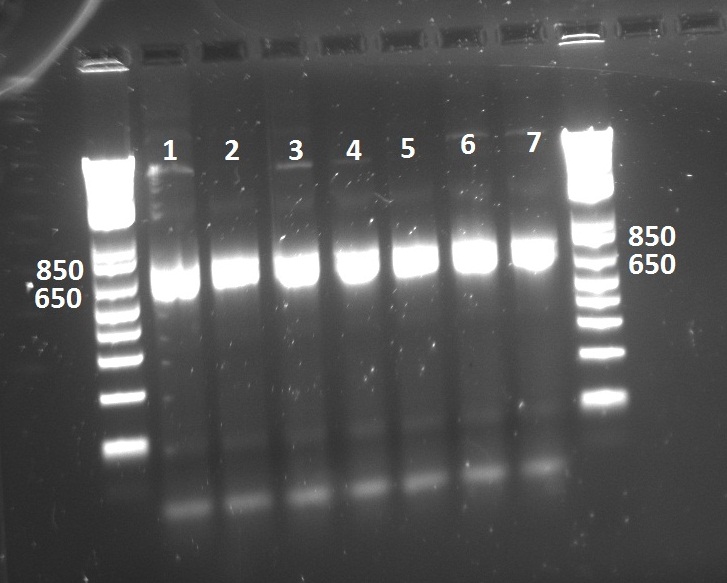
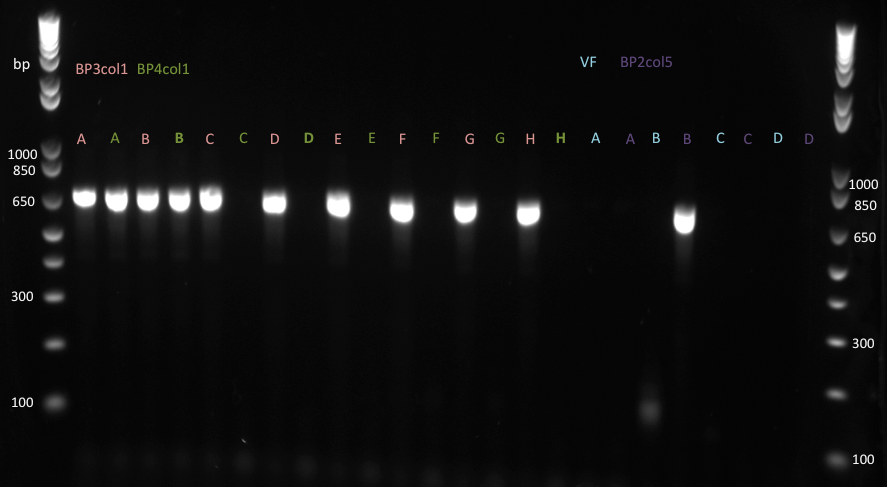
Lilia tested the expression of the TetR mutants with the T7 promoter. The expression mix contained the Green Lysine and the gel fluorescence was imaged and is shown on the next picture.
The BP2col2 does not express, what explains the failure of the protein pool down at the MITOMI made on Wednesday.
Friday, September 2 2011
Alina wanted to try a new extension PCR of the T4-lysis and a Gibson of the exteneded-lysis into the K1 backbone. The transformation was successful with tons of colonies. A colony PCR will be run on Monday.
Nadine came back in the lab and cotransformed pSB3K1 Pconst-TetR with J61002 Plac-RFP since the sequencing results were concluent, in order to perform IPTG and ATC platereader experiments.
We had a lot of contaminated bottles, so she also made new SOC bottles.
Monday, September 5 2011
Alina ran a colony PCR on the K1-T7-lysis Gibson assembly.
After having set liquid cultures on Saturday, Nadine set up the platereader experiment as follows:
- J61002 Plac-RFP with increasing concentrations of IPTG
- pSB3K1 TetR-LacI + J61002 Plac-RFP with increasing concentrations of IPTG
- pSB3K1 TetR-LacI + J61002 Plac-RFP with increasing concentrations of ATC
She had also prepared co-transformations with pSB3K1 TetR-LacI + J61002 Plac-lysis, but the sequencing results indicated that the lysis cassette was wrong.
Tuesday, 6 September 2011
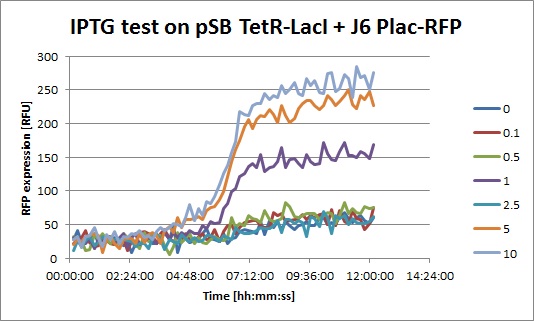
The results from the platereader experiment came back and they look good!
- J6 Plac-RFP: The level of RFP expression doesn't rise much with high doses of IPTG, showing that DH5alpha cells don't express much LacI and that Plac is a quite weak promoter.
- J6 Plac-RFP + pSB TetR-LacI, IPTG: you can clearly see an increase of RFP when IPTG is added. This means that even with a constitutive promoter in front of TetR, LacI is still expressed. In fact, Pconst is weaker than Ptet. The "2.5" data seems strange, I perhaps missed to put IPTG in this well.
- J6 Plac-RFP + pSB TetR-LacI, ATC: Without ATC, we have a similar expression curve as in the IPTG experiment. When ATC is added, we see a diminution of RFP expression because LacI is not repressed anymore.
Vincent transformed the "randomer" Gibson assemblies (from nearly a month ago) into BL21 cells. If you cross-check with the numerical scheme, those would be variants 7, 8, 9, 16, 17, 18.
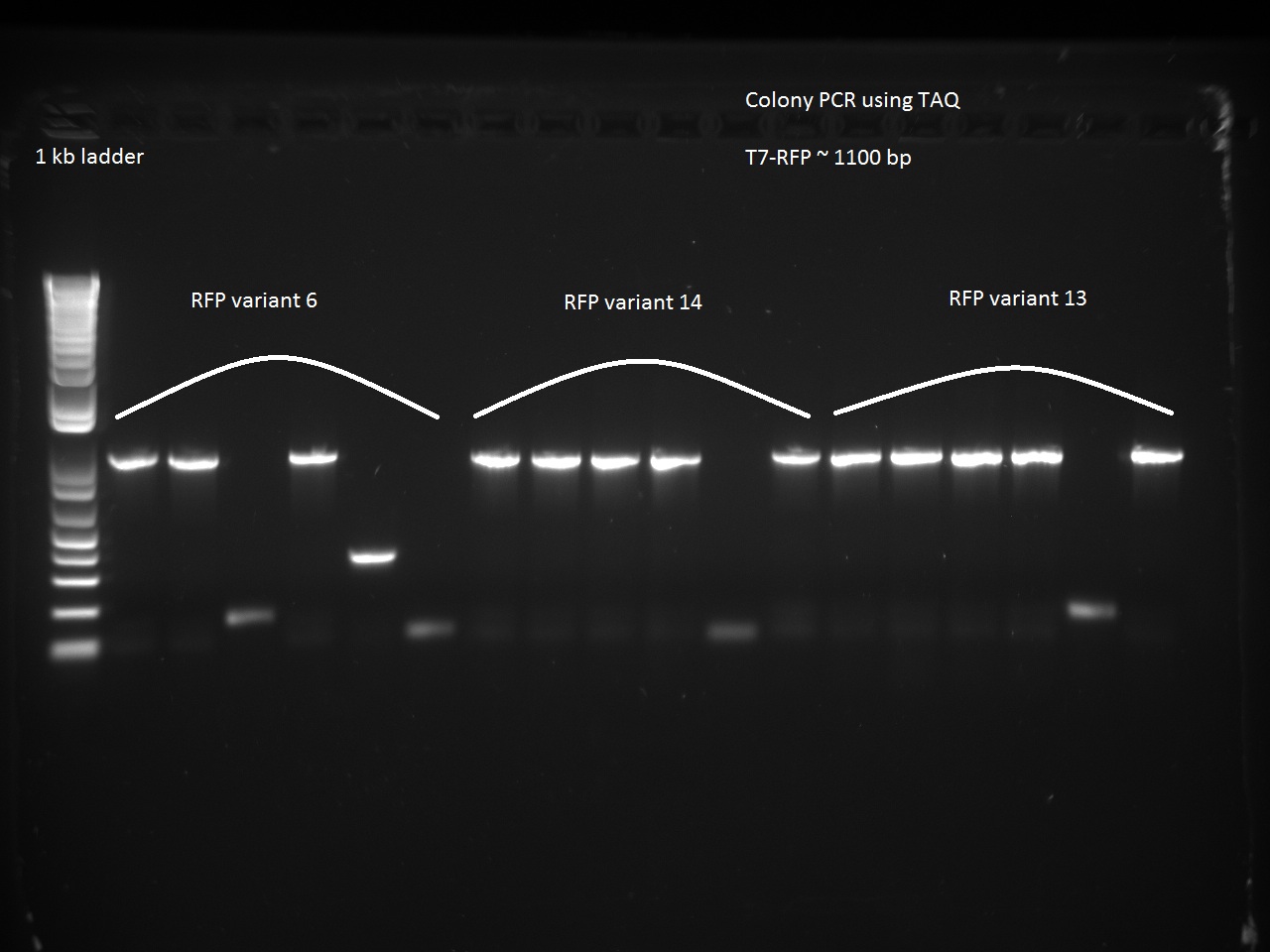
We also went after the remaining non-randomer variants that were left (6, 13, and 14) using a colony PCR on six colonies of each variant. The PCR used 64 C for annealing and 1 minute for extension.
Wednesday, 7 September 2011
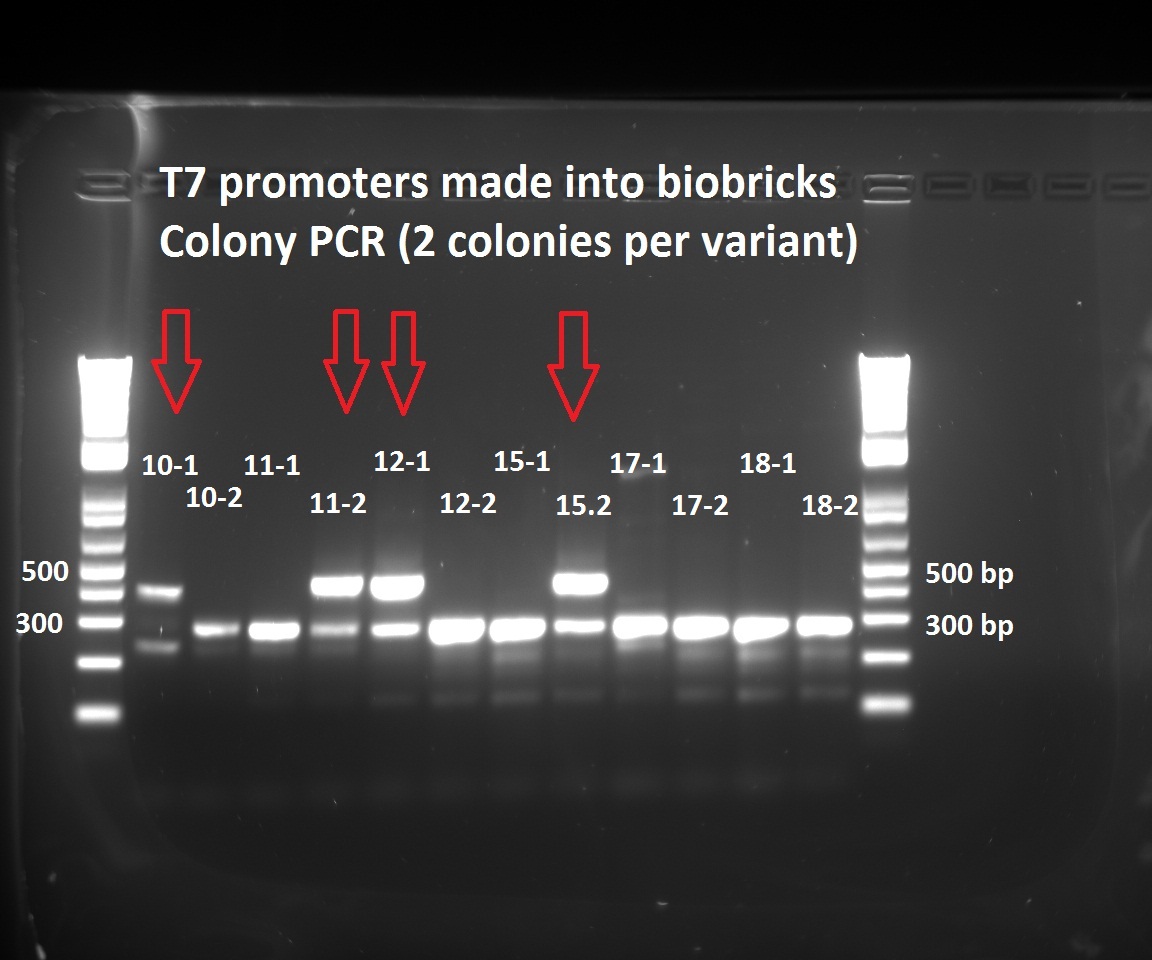
Nadine made colony PCRs for the T7 promoter variants that Henrike cloned into the biobrick vector. The right insert should be ~450 bp long, but all the colonies show a fragment at 300 bp. Nevertheless, we have 5 colonies with the 450 insert. The low number of positive colonies correlates with the observation that the ligation negative control plate had about half as much colonies as the different ligations.
She also prepared some liquid cultures for tomorrow's platereader experiment, and miniprepped the pSB TetR-LacI plasmid before sending it for 2 new sequencing reactions.
Vdog made glycerol stocks and minipreps of the non-randomer RFP variants 6, 13, and 14 as well as Alina's T7-const-C2/C11, T7 lac 1, and T7 lac2 3.
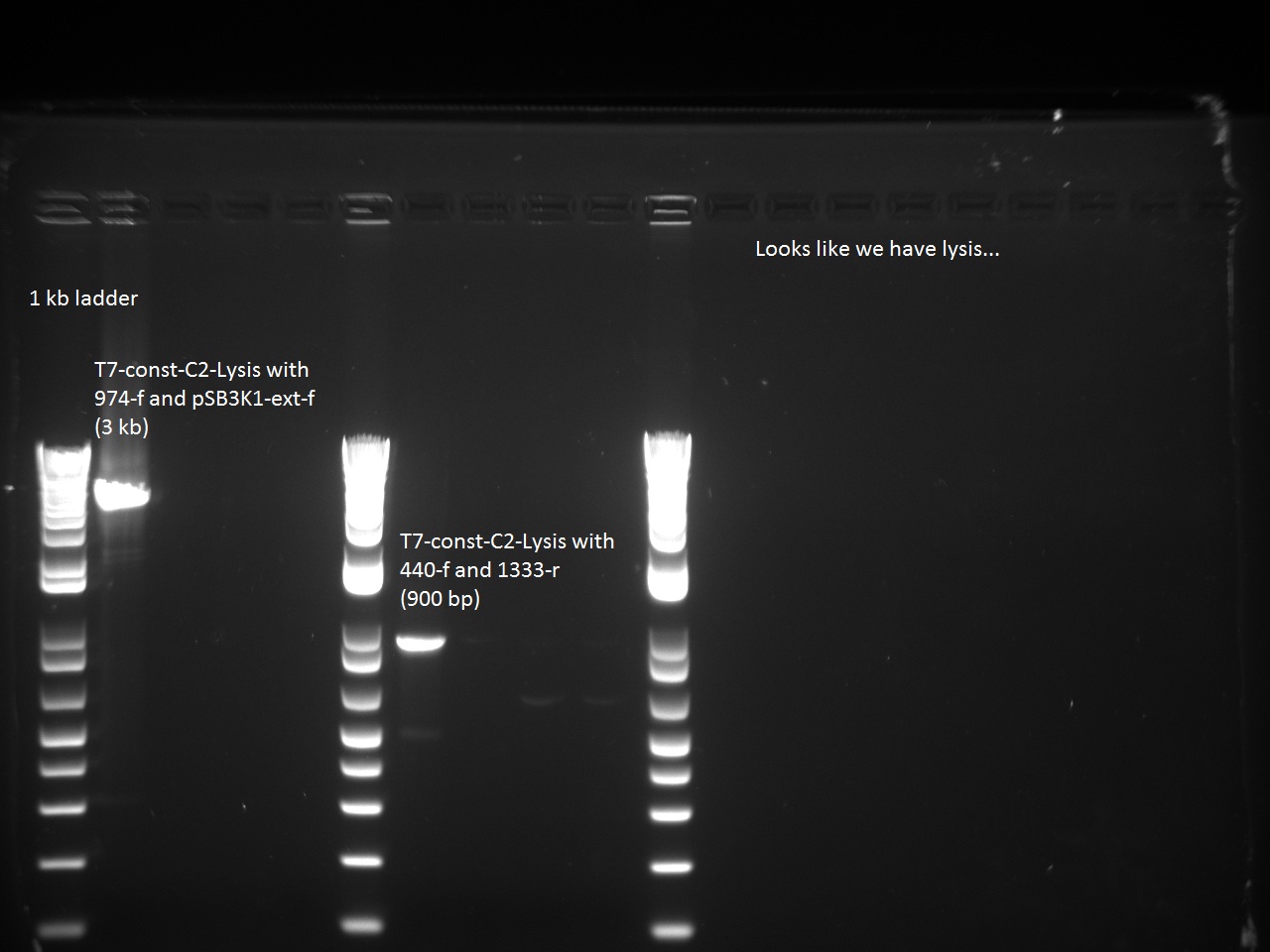
He also sent all of Alina's constructs (aforementioned) for sequencing using two different primers: 947_f and 816_r. He then ran a control PCR on all the T7 constructs using Alina's EPFL TAQ with the following combination of primers:
- 440-f and 1333-r
- 947-f and pSB3K1-ext-r
The protocol asked for a 25 uL aliquot with
- 2.5 uL Thermo Pol buffer (TPB)
- 0.5 uL dNTP
- 0.5 uL primer 1
- 0.5 uL primer 2
- 1 uL plasmid
- 0.25 uL TAQ
He went on to transform the non-randomer RFP minipreps and the T7-const-c2/c11, lac1, and lac2 3 constructs into BL21 cells This was followed by the production of a 96 round-bottom well plate with 100 uL of LB with 1 uL of Kan. Twelve colonies from each plate were chosen and swirled into the individual wells. This plate was put on a shaker overnight (after having been sealed with the Easy-Breathe adhesive paper).
Thursday, 8 September 2011
Sequencing of T7-const-C2 returned the correct sequence, as did the Control PCR, so it seems that the lysis cassette has found its way into the T7-const-C2!
On the other hand, sequencing results for pSB TetR-LacI show that the Ptet promoter before LacI is mutated: we have 3 deletions and one mismatch. The deletions are in the two binding sites of TetR, probably preventing TetR action on the promoter. This can then explain why we had a low RFP expression in the paltereader for the pSB TetR-LacI + J6 Plac-RFP cotransformation: we thought Pconst was too weak, leading to a too big LacI expression, but actually the problem was that TetR couldn't repress well LacI.
Nadine made liquid cultures of J61002 Ptet-RFP because we were running out of DNA. We want to send it for sequencing, to be sure that in this plasmid Ptet is OK - although the experiments suggest that yes. She also made liquid cultures of the successful ligations (see yesterday's results) in order to sequence-verify them afterwards.
Vincent transformed T7-const-C2-Lysis and T7-const-C11-Lysis (as a negative control) into BL21 cells and plated them. He also co-transformed:
- T7-const-C2-Lysis + Ptet-GFP
- T7-const-C2-Lysis + Ptet-RFP
- T7-const-C11-Lysis + Ptet-GFP
- T7-const-C11-Lysis + Ptet-RFP
into BL21 using Amp + Kan plates. He used 1 uL of each plasmid.
Vince also prepared a 96 well plate for testing all the randomers (7, 8, 9, 16, 17, 18) of T7-RFP and grew it overnight (shaking, with Easy-Breathe paper to let the cells get air).
Friday, 09 September 2011
Nadine's platereader experiment gave disappointing results: the cells didn't grow much, reaching only 0.1-0.2 OD instead of 1 normally. This will be repeated next Tuesday. The minipreps of J6 Ptet-RFP and the T7 biobricks yielded high concentrations, and therefore were sent for sequencing.
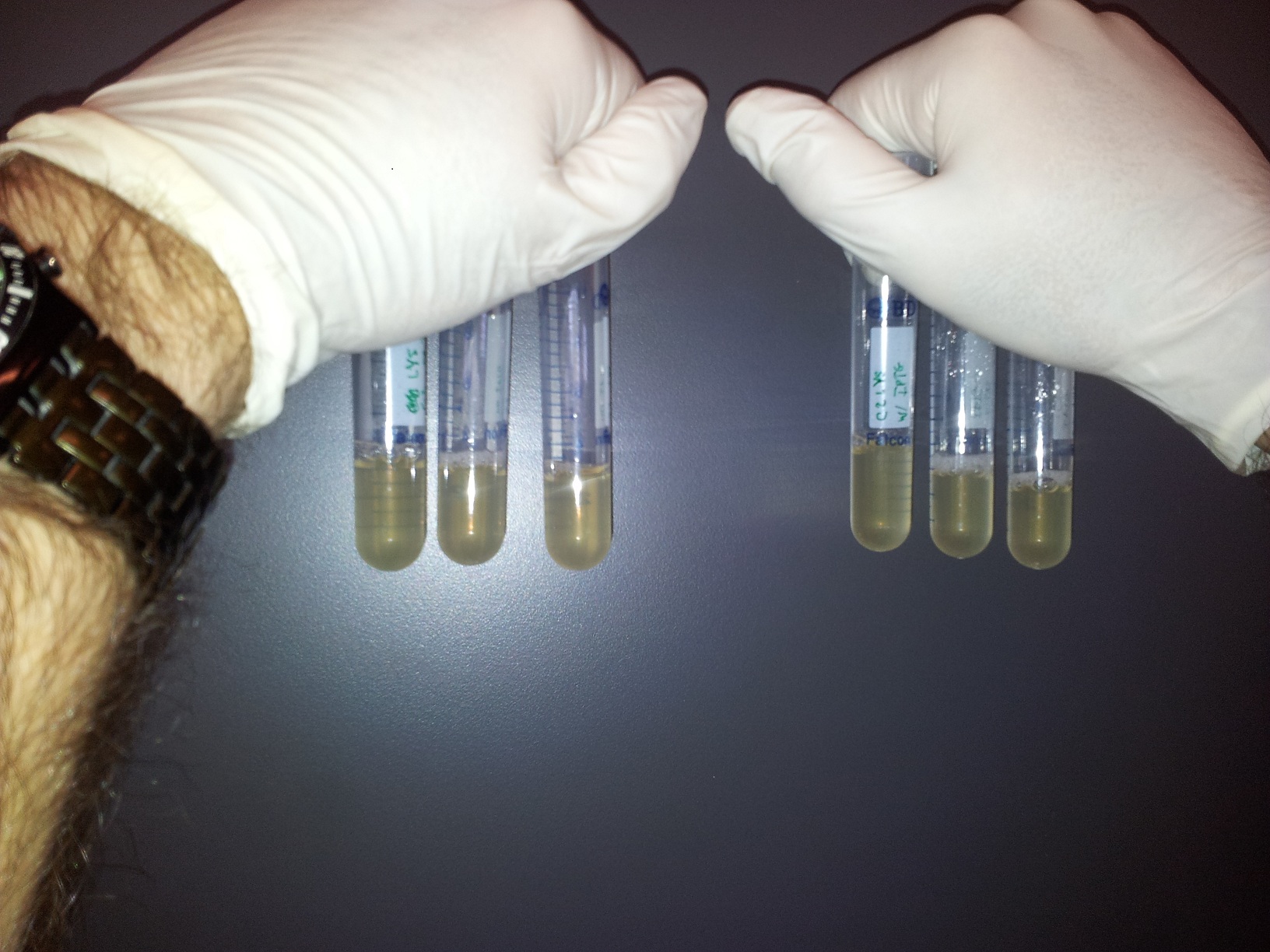
Vince made 2 new plates from his "main" plate that had grown overnight, and he grew those plates to make them into glycerol stocks. He took the "main" plate to the Bioscreening Facility to use their platereader. Meanwhile, he set up a time-lapse, qualitative test of the lysing ability of the C2 culture. We set up two sets of three test tubes, one with C2 induced with 250 uM IPTG and the other un-induced. Here are the photos, and, as you can perhaps see, the last one shows some lysis.
The randomer plate-reader experiment went well and results should be appearing shortly. We used 250 uM IPTG. Vincent made liquid cultures of all 12 of the T7 and T7-lac2 RFP variants for a big plate-reader experiment on Saturday.
Saturday, 10 September 2011
Vincent ran the plate-reader experiment for the 12 T7, T7-lac2 RFP variants, using 250 uM IPTG and 0 uM as a negative control. The induction curves look great, results should be coming in shortly.
Vdog made liquid cultures of T7-C2, T7-C11 and T7-RFP-1 for the lysis platereader experiment.
Sunday, 11 September 2011
Vincent ran another plate-reader experiment, this time for lysis. We tested three different IPTG concentrations: 0 uM, 100 uM, 250 uM, and 500 uM. Two different types of dilution were used: 1:10 and 1:20 (to see how the initial dilution affected the lysis curves). Finally, we checked to see how C2 behaved versus C11 (which does not have the lysis cassette but has T7-const) and versus T7-RFP (as far as induction time is concerned). Results should be appearing shortly.
Someday, 12 September 2011
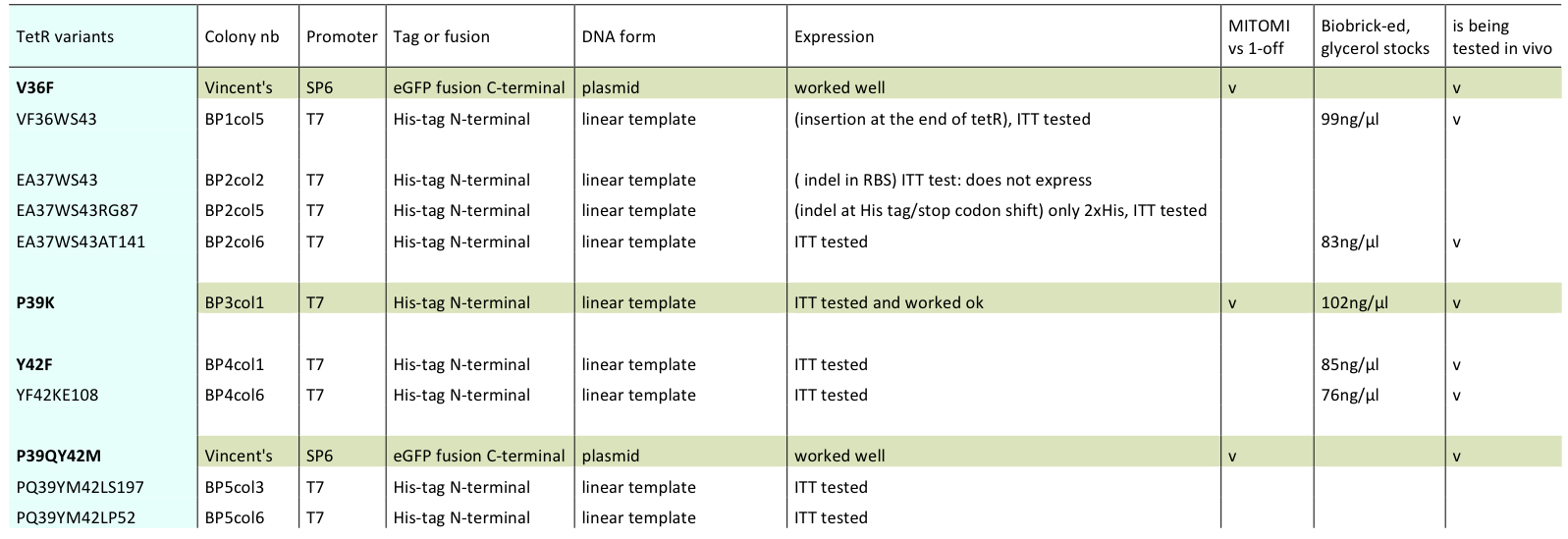
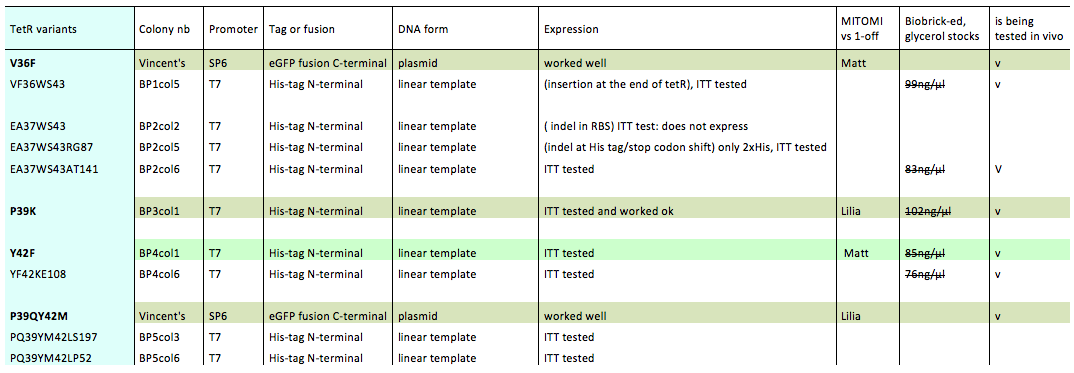
Lilia prepared minipreps of the 5 muTetRs in Biobrick standart (pSB3K1), they will be send for sequencing. Here is the updated list of available mutants that are being characterized:
Nadine started liquid cultures for tomorrow's platereader experiment. She also made the PCRs for putting TetR mutants into the pSB3K1 Pconst-TetR plasmid by Gibson. We can see the expected band at around 700 bp! Unfortunately, amplification of the pSB3K1 backbone didn't work...
The mutants used were:
- BP1col5 (V36FW43S)
- BP2col6 (E37AW43SA141
- BP3col1 (P39K)
- BP4col1 (Y42F)
- BP4col6 (Y42FK108E)
- V36F
- P39QY42M
Tuesday, 13 September 2011
Nadine re-did the PCR to amplify pSB3K1 backbone, this time using the right template DNA and it worked! She had strange results on the platereader experiment again, but it's because she diluted cells in water instead of LB medium. The experiment will be re-ran a 3rd time!
About the T7 biobricks, the sequencing results show that we already have T7 const (1) and T7 Lac2 030 (12). The other most interesting promoter variants are: T7 030 (3), T7 054 (4), T7 111 (6), T7 lac2 054 (13) and T7 lac2 111 (15). We will focus on them and try to build biobrick formats before the deadline.
Wednesday, 14 September 2011
Nadine tried to purify the PCR products of the TetR mutants, but lost them during the process... So she re-ran the PCR and used it directly do do Gibson assemblies that she later transformed into DH5alpha cells.
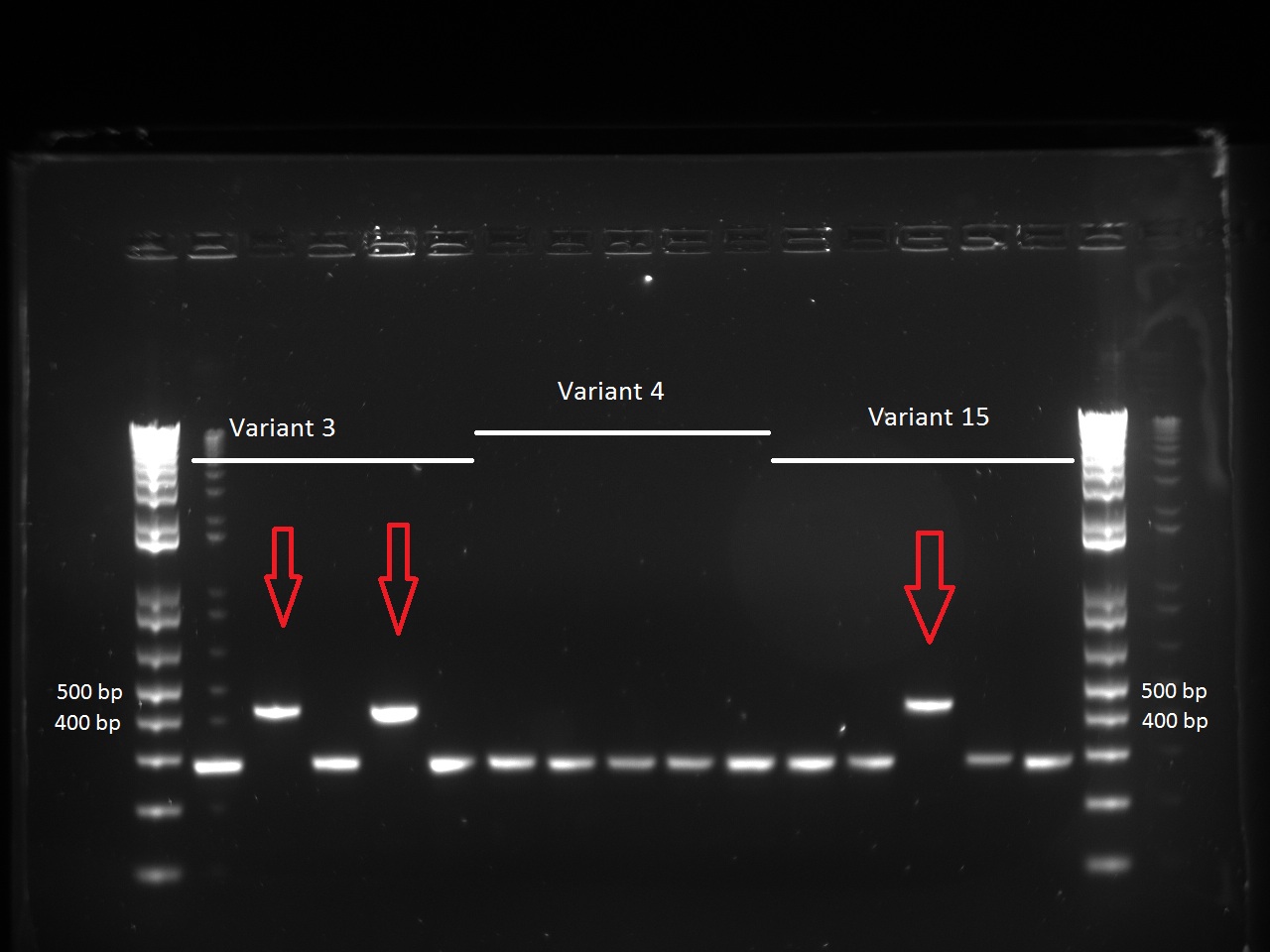
She looked at the plates containing the biobrick variants and made colony PCRs on five colonies of variats 3,4 and 15. The resuklts show that we have 2 good colonies for 3 and one for 5.
Finally, the Plac promoter being correctly inserted into J61002 Plac-RFP plasmid, Nadine also ran a PCR to add biobrick extensions to the promoter.
Vdog ran a ten-hour experiment with the help of Henrike and Matt. We made two 100 mL batches LB with Kan and Amp in 1:1000 dilutions. We then took the two sets of four tubes of 5 mL liquid cultures (of C2-Lys-Ptet-RFP & C11-Lys-Ptet-RFP) and spun them down, threw away the supernatant, resuspended them in 5 mL of PBS buffer, spun them down again, resuspended once more in 5 mL of PBS buffer, and then spun them down again before resuspending one last time in 1 mL of PBS buffer. The final tube of resuspended and clean culture contained no more than 4 mL. We then took cuvettes and measured the OD of each culture in a 1:100 dilution. We then calculated the amount of liquid culture we would need to put in each flask to produce an equal starting OD for both flasks of around .4
Every hour, we took 1 mL samples from both flasks, spun them down, sterile filtered the supernatant, and stored the resulting filtered supernatant for transformations the next day. We also took OD measurements of samples from the flask. The flasks were incubated for an hour at 37 C before being taken out for measurements.
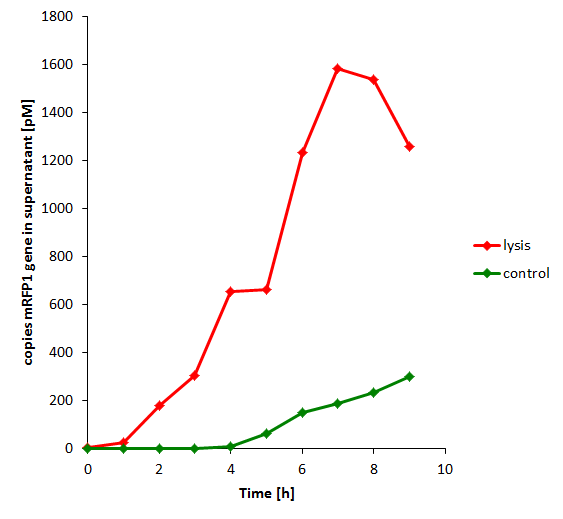
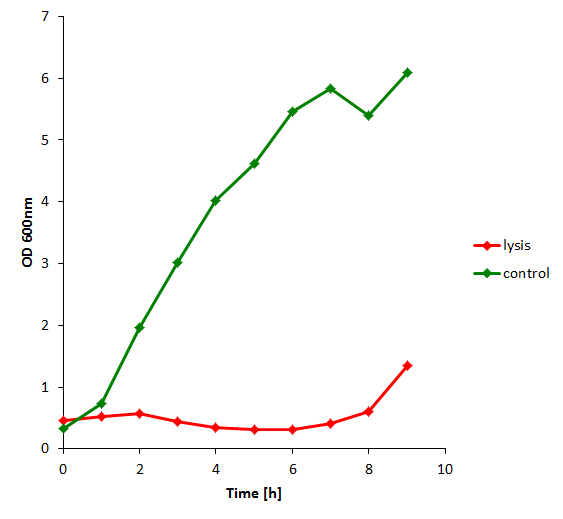
The results of the experiment indicate that not only do we have successful lysing of the relevant cells, but we also are able to recover the relevant DNA, not only as is found in the supernatant by qPCR but also by looking at the number of colonies produced by each hourly sample when transformed.
Thursday, 15 September 2011
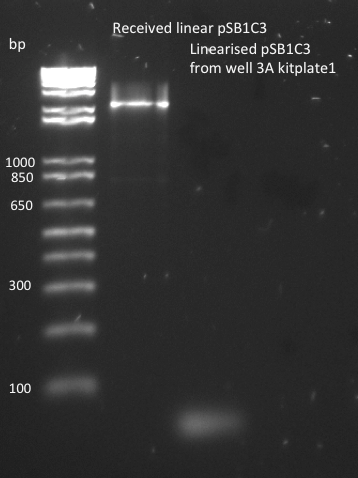
Lilia prepared pSB1C3 backbone from distribution KitPlate in linearised form and also transformed it DH5alpha. If the transformation works, minipreps on pSB1C3 will be done tomorrow.
4x1L of LB-agar were prepared and should be autoclaved by tomorrow in AI. LB-kanamycin plates are ready and will be brought to BM.
Nadine tested 5 colonies on each Gibson assembly plate, but with no results... So Gibson need to be done again, this time purifying the TetRs before. She also digested the Plac biobrick fragment with EcoRI and PstI.
Friday, 16 September 2011
Lilia made colony PCR on muTetR variants from the 2nd batch of biobricking: TetR_Biobrick_f &_n primers and EPFL taq polymerase were used.
The same PCR on plasmids from the first batch of biobricked mutants gave an unexpected band at higher MW than what was expected. Same for each of five mutants.
The pSB1C3 biobrick backbone was linearized by PCR with the pSB_prep_2Eb and pSB_prep_3P-1 primers, but the PCR failed - there is no plasmid band in the second well on the gel. It was also transformed into DH5alpha on CM LB-agar plate, but that failed because pSB1C3 in incompatible with DH5alpha cells.
Nadine ligated the Plac biobrick into pSB1C3 and transformed. She also purified the TetR genes with Gibson extensions, sending them for sequencing after having done a new Gibson reaction. The Gibson reactions were also transformed, hoping that this time it will work.
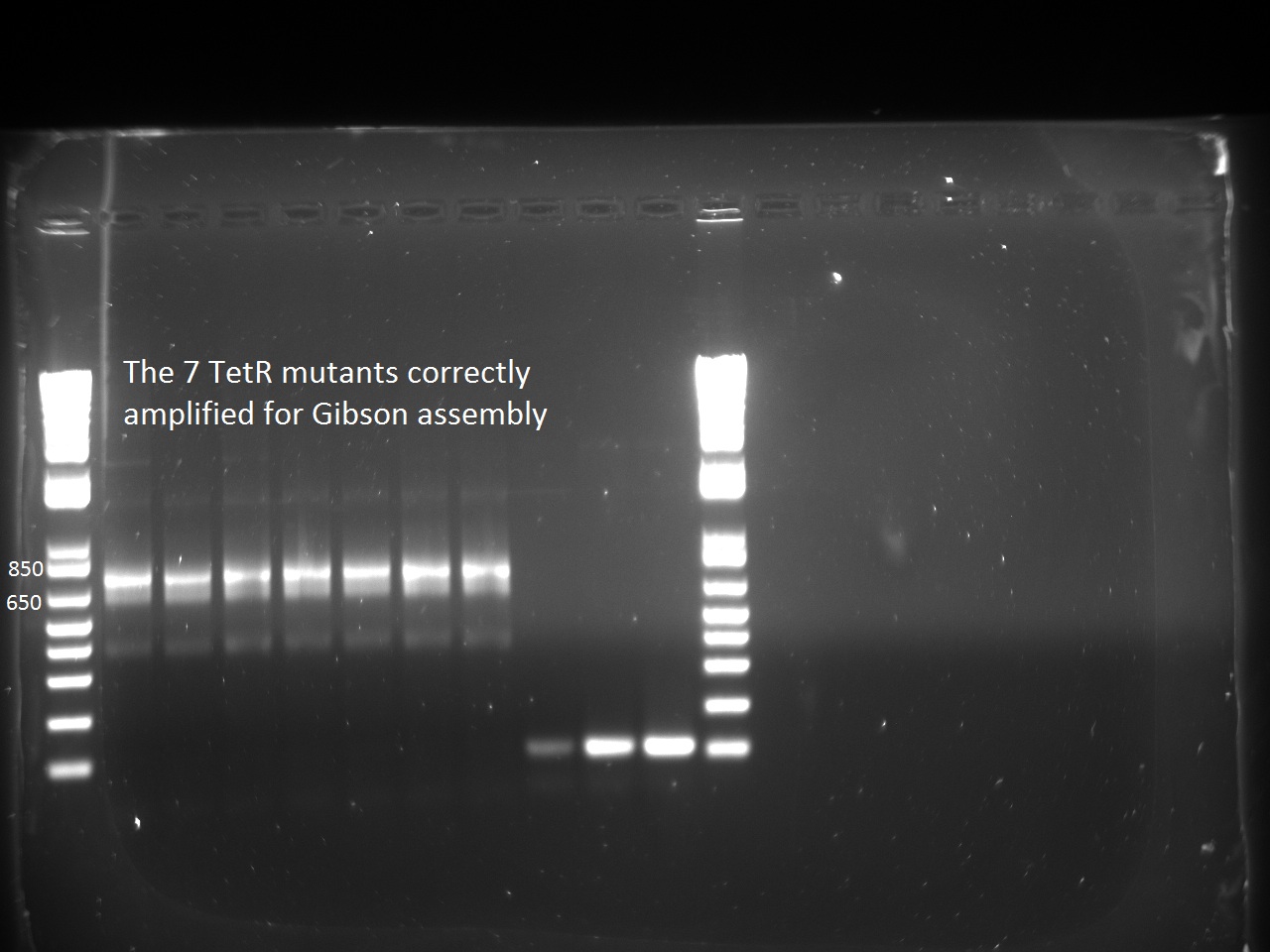
Second attempt to PCR tetR mutants with BioBrick primers yield in PCR product for all the mutants we have:
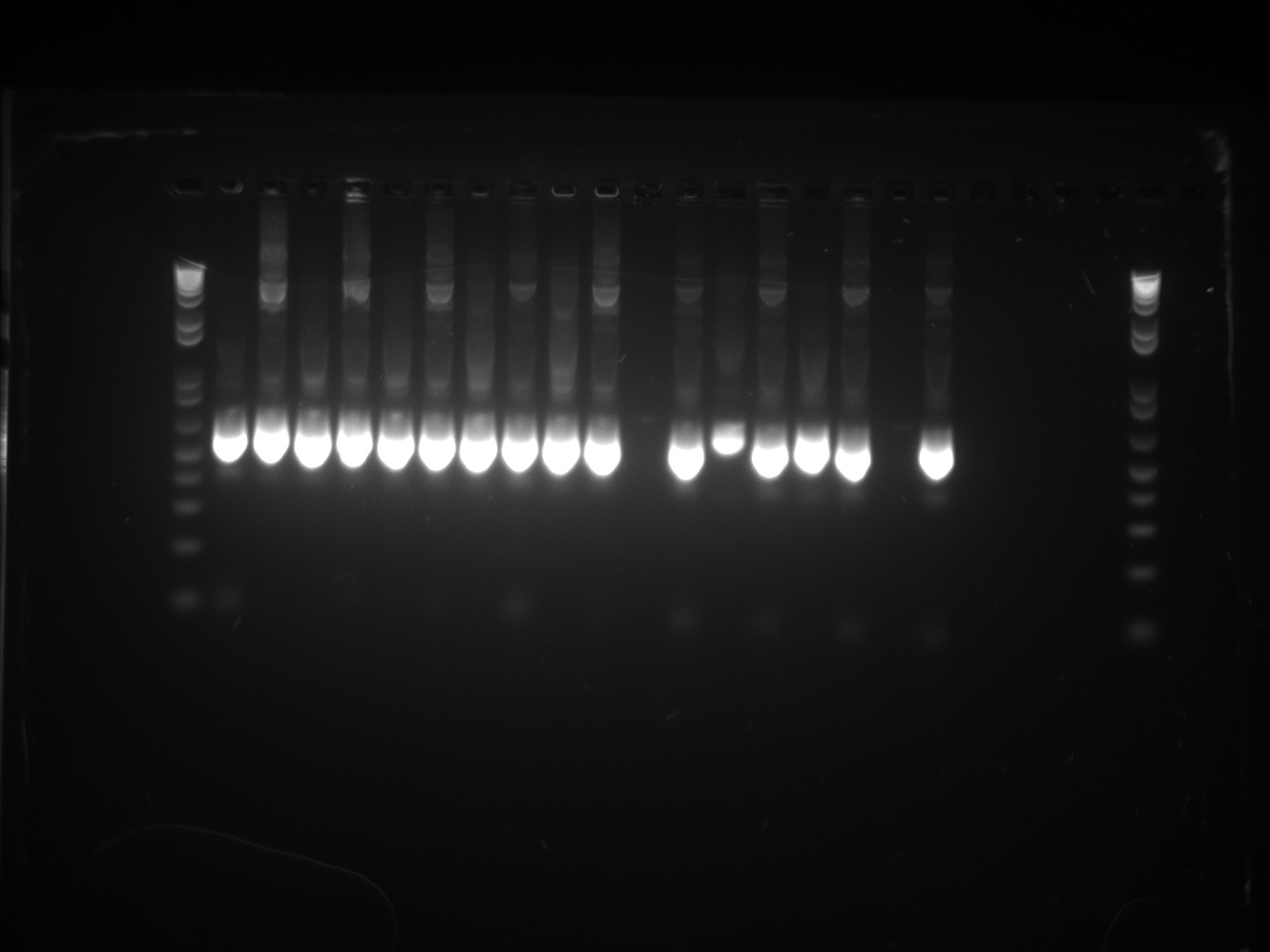
We will now digest and ligate it to the pSB1C3 backbone!
Saturday, 17 September 2011
Nadine made colony PCRs on the Gibson assemblies with TetR mutants and on Plac-biobrick ligations. For the Plac-biobrick, all the 5 colonies tested seem to have the Plac insert.
Doug made all the graphs of Nadine's platereader experiments. We still have some platereaders left to do: J61002 Ptet-RFP wil be done tomorrow and hopefully the TetR mutants will be ready and co-transformed on Tuesday.
 "
"