Team:EPF-Lausanne/Protocols/Gibson assembly
From 2011.igem.org
(Difference between revisions)
(Created page with " Protocol adapted from Nikolaus Obholzer (https://wiki.med.harvard.edu/SysBio/Megason/IsothermalAssembly) * Take the different DNA parts you want to assemble and measure their c...") |
|||
| (7 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:EPF-Lausanne/Templates/ProtocolHeader|title=Gibson assembly}} | ||
| + | |||
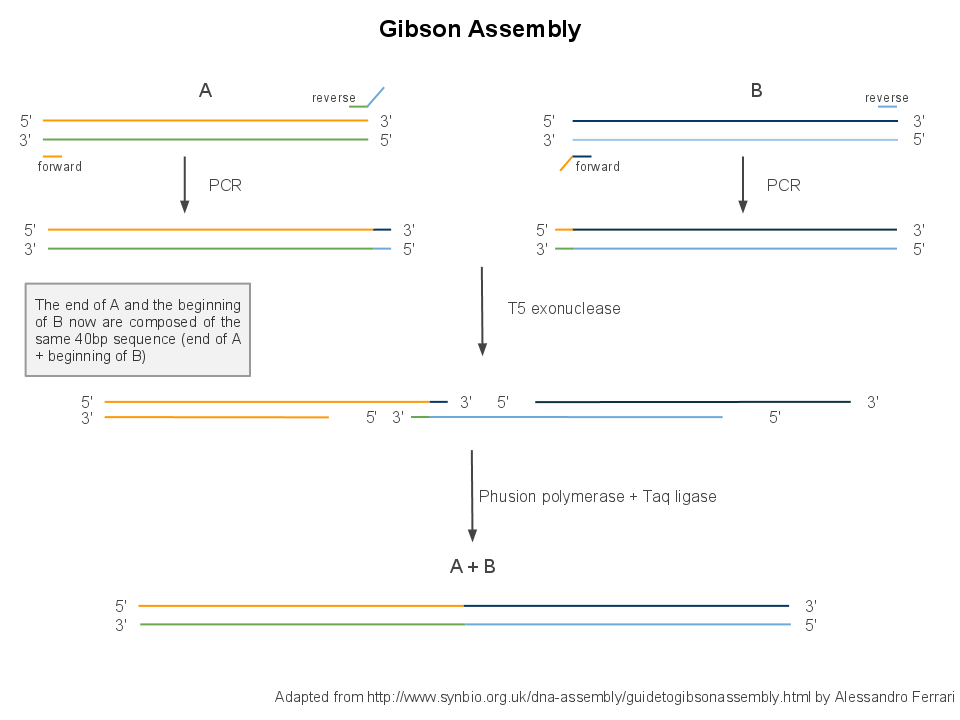
| + | We use Gibson assembly to assemble DNA fragments into plasmids. It is a multistep process, requiring first a PCR to create each fragment by copying them out of template plasmids, then a single isothermal assembly to stick the fragments together, then transformation into competent cells and culture for amplification. | ||
| + | |||
| + | [[File:EPFL2011 GibsonAssemblystrandscoloured.png|700px]] | ||
Protocol adapted from Nikolaus Obholzer (https://wiki.med.harvard.edu/SysBio/Megason/IsothermalAssembly) | Protocol adapted from Nikolaus Obholzer (https://wiki.med.harvard.edu/SysBio/Megason/IsothermalAssembly) | ||
| Line 6: | Line 11: | ||
* Based on these numbers, calculate the ul you need of each to have the same amount of molecules | * Based on these numbers, calculate the ul you need of each to have the same amount of molecules | ||
* Take 5 ul of the mixed solution and add 15 ul of Gibson mastermix | * Take 5 ul of the mixed solution and add 15 ul of Gibson mastermix | ||
| - | * Heat at | + | * Heat at 50°C for 45-60 minutes (there is a programm written in the iGEM folder) |
| - | + | ||
| - | + | Now the samples are ready to be transformed. | |
PS: if someone has a less complicated method, please change the protocol :) | PS: if someone has a less complicated method, please change the protocol :) | ||
| + | |||
| + | |||
| + | {{:Team:EPF-Lausanne/Templates/Footer}} | ||
Latest revision as of 16:05, 2 September 2011
Gibson assembly
Back to protocols.We use Gibson assembly to assemble DNA fragments into plasmids. It is a multistep process, requiring first a PCR to create each fragment by copying them out of template plasmids, then a single isothermal assembly to stick the fragments together, then transformation into competent cells and culture for amplification.
Protocol adapted from Nikolaus Obholzer (https://wiki.med.harvard.edu/SysBio/Megason/IsothermalAssembly)
- Take the different DNA parts you want to assemble and measure their concentration with Nanodrop
- We need equal molecuar ratios! Divide the concentration measured above [ng/ul] by the length of each part to have equivalent numbers
- Based on these numbers, calculate the ul you need of each to have the same amount of molecules
- Take 5 ul of the mixed solution and add 15 ul of Gibson mastermix
- Heat at 50°C for 45-60 minutes (there is a programm written in the iGEM folder)
Now the samples are ready to be transformed.
PS: if someone has a less complicated method, please change the protocol :)
 "
"